Bdellovibrio
A Microbial Biorealm page on the genus Bdellovibrio
Classification
Higher order taxa
Kingdom: Bacteria Phylum: Proteobacteria Class: Deltaproteobacteria Order: Bdellovibrionales Family: Bdellovibrionaceae Genus: Bdellovibrio
Species: Bdellovibrio bacteriovorus

|
NCBI: Taxonomy |
Description and significance
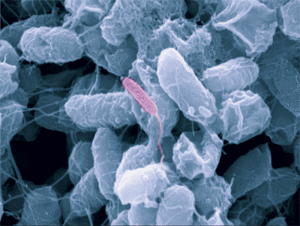
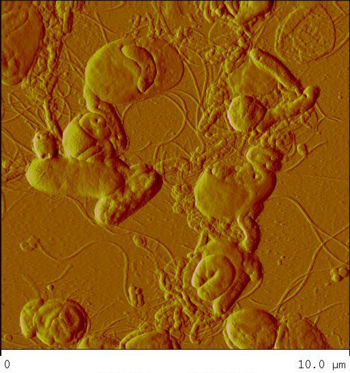
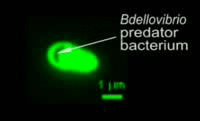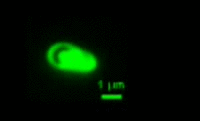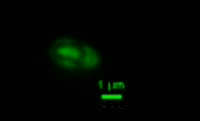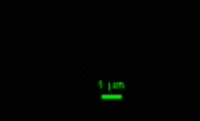
Bdellovibrio is a genus of small, highly motile, vibrio shaped, monoflagellated, gram negative bacteria of the class delta-proteobacteria with the ability to parasitize and kill other gram negative bacteria[1]. This includes many pathogens, a few of the genera being: Alcaligenes, Campylobacter, Erwinia, Escherichia, Helicobacter, Pseudomonas, Salmonella, Legionella, and Shigella[1]. Bdellovibrio have what is described as a biphasic lifestyle with a motile, free-living “hunting” phase and an intra-cellular parasitizing growth phase [1],[2]. The intra-cellular phase of Bdellovibrio is restricted to the periplasm, where the prey’s physiology is modified to suit the predator, forming a distinctive bdelloplast[5]. Wild type Bdellovibrio are obligate parasites and must live out part of their lives in a host or die[4]; however, researchers have been able to artificially culture certain strains of Bdellovibrio on rich media in the lab. Bdellovibrio are normally grown in E. coli prey cultures in the lab, and colonies are isolated from plaques in confluent lawns of prey culture, similar to viruses[2]. The most widespread species, Bdellovibrio Bacteriovorus, inhabits a wide range of environments such as freshwater, brackish water, seawater, sewage, water pipes, animal intestines and other water reservoirs[3],[8].
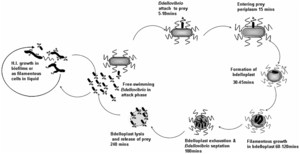
This bacterium is also commonly found in man-made habitats, the rhizosphere of plant roots, and soil. It has also been observed to reside in the intestinal tract of certain mammals where it reduces pathogens in the intestines. B. bacteriovorus is useful in the purification of waste water because it decreases gram-negative bacterial counts.
The life cycle of B. bacteriovorus includes five stages. First, the bacterium attaches itself to the host's outer membrane using a hook-like extension which breaks down the outer cell wall through the release of certain enzymes. This is followed by penetration into the periplasm and the repair of damage done to the cell membrane. Once the bacterium is inside the periplasm, it introduces hydrolytic enzymes into the host cytoplasm. This increases the bacterium’s food supply by making the inner membrane of the host cell “leaky.” The bacterium then grows and once it has exhausted all the resources in the host’s cell, it divides into as many as 15 motile cells. The weakened host cell lyses, releasing progeny B. bacteriovirus bacteria, which are then ready to find new prey to attack. This life cycle takes up to three to four hours.
Genome structure
The genome of B. bacterivorus has been recently published in 2004 by Rendulic et al. and has revealed a number of characteristics of, and mechanisms employed in its predatory lifestyle[4]. After initial discovery by Hans Stolp in 1963 and a period of enthusiastic biochemical and physiological research, the effort substantially dropped. It wasn’t until recently with the advent of molecular genetics and sequencing that work on Bdellovibrio has picked up[6]. Sequencing of the HD(Host-Dependent)100 strain yielded a large 3.85Mb genome which was surprising considering the small size of the bacterium (0.2 to 0.5 μm by 0.5 to 2.5 μm)[4],[6]. The genome is devoid of any plasmids and contains only a single IS element and one prophage[4]. The G/C content of the genome was also fairly constant among all genes which indicates that no lateral gene transfers have occurred in recent times[7], despite being in constant contact with prey genomes containing high G/C content[4],[6].
Predatosome and Specialization
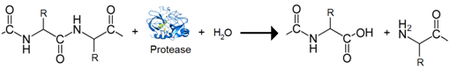
The Bdellovibrio genome is of comparable size to many non-predatory bacteria; however, its composition is vastly different. The largest portion is dedicated to an essential group of proteins that constitute the heart of its predatory nature. The predatosome constitutes 40% of the B. bacteriovorus genome and includes genes that have roles in killing and digesting the prey, and regulatory genes that control the processes[2]. With the exception of a single endosymbiont possessing a tiny genome, B. bacteriovorus has the highest density of protease and peptidase genes reported[4]. The predatosome includes 20 DNAses, 9 RNAses, 15 lipases, 10 glycanases, over 150 protease and peptidases, and 89 other hydrolases. This is in stark contrast to a non-predatory bacterium such as E. coli, with 3 DNAses, 8 RNAses, 2 lipases, and 60 proteases/peptidases mainly used for cellular remodeling[6]. The numerous DNAses are a likely explanation for the lack of lateral gene transfer observed, as the prey genetic material is fully degraded leaving no genes intact to undergo transfer[7]. Furthermore, B. bacteriovorus possesses only 11 of the essential amino acid synthesis pathways and is missing 10 amino acid degradation pathways[4]. This dedication to the predatosome, as well as the very condensed and non-permissive nature of its genome towards other genetic elements indicates a high level of specialization and is consistent with Bdellovibrio’s obligate parasitic lifestyle.
Biocontrol and therapeutic Agents
The ability of Bdellovibrio to lyse bacterial cells while growing in a non-competitive inter-periplasmic niche has made them potential candidates as live biocontrol, and perhaps even therapeutic agents[1],[8],[9],[10]. Data from the recent genome publication by Rendulic et. al has indicated an absence of a type III secretion system that is heavily associated with pathogenic infection of eukaryotic organisms, such as humans, in B. bacteriovorus[4],[11]. There are also no reported incidents of Bdellovibrio bacteria invading mammalian cells[4]. These are positive signs for the possible use of Bdellovibrio as therapeutic agents in animals and possibly in humans.
Ongoing Trials
Atterbury et al. have already conducted trials testing B. Bacteriovorus use in eradicating Salmonella enteritidis in young chicks[10]. The birds showed no side effects and had significantly reduced salmonella numbers when administered an oral dose of predatory HD100 Bdellovibrio, as opposed to the control or a genetically modified non-predatory Bdellovibrio strain[10]. B. bacteriovorus was also able to significantly reduce the biomass of E. coli and Pseudomonas fluorescens biofilms formed on microtiter plates[9]. These are important applications with very extensive economical, health, and ecological implications. For example in 2010, there was an enormous recall of over half a billion eggs contaminated with Salmonella enteritidis[10]. Moreover, the use of B. bacteriovorus can eliminate the need for extensive antibiotic therapies, especially in feed for livestock, which promotes antibiotic resistance[10]. It can also reduce the need for harmful chemicals used to clear strongly adhering biofilms[9]. The possibilities of applications of B. bacteriovorus are endless, but more research must be conducted to affirm the safety and reliability of such use.
References
1Markelova N. “Predacious Bacteria, Bdellovibrio With Potential for Biocontrol.” International Journal of Hygiene and Environmental Health, 2010, DOI: 10.1016/j.ijheh.2010.08.004
2Lambert C., Chang C., Capeness M., and Sockett E. “The First Bite— Profiling the Predatosome in the Bacterial Pathogen Bdellovibrio,” PLoS One, 2010, DOI: 10.1371/journal.pone.0008599
3Chu W. and Zhu W. “Isolation of Bdellovibrio as Biological Therapeutic Agents Used for the Treatment of Aeromonas hydrophila Infection in Fish.” Zoonoses and Public Health, 2010, DOI: 10.1111/j.1863-2378.2008.01224.x
4Rendulic S., Goesmann A., Meyer F., Sockett E., Schuster S., Jagtap P., Rosinus A., Eppinger M., Baar C., Lanz C., Keller H., Lambert C., Evans K. “A Predator Unmasked: Life Cycle of Bdellovibrio bacteriovorus from a Genomic Perspective.” Science, 2004, DOI: 10.1126/science.1093027
5Lambert C., Hobley L., Chang C., Fenton A., Capeness M., Sockett L. “A Predatory Patchwork: Membrane and Surface Structures of Bdellovibrio bacteriovorus.” Advances in Microbial Physiology, 2008, DOI: 10.1016/S0065-2911(08)00005-2
6Sockett E. “Predatory Lifestyle of Bdellovibrio bacteriovorus.” Annual Review of Microbiology, 2009, DOI: 10.1146/annurev.micro.091208.073346
7Gophnaa U., Charleboisb R., Doolittle F. “Ancient Lateral Gene Transfer in the Evolution of Bdellovibrio bacteriovorus.” Trends in Microbiology, 2006, DOI: 10.1016/j.tim.2005.12.008
8Sockett E. and Lambert C. “Bdellovibrio as therapeutic agents: a predatory renaissance?” Nature Reviews Microbiology, 2004, DOI: 10.1038/nrmicro959
9Kadouri D. and O'Toole G. “Susceptibility of Biofilms to Bdellovibrio bacteriovorus Attack.” Applied and Environmental Microbiology, 2005, DOI : 10.1128/AEM.71.7.4044-4051.2005
10Atterbury R., Hobley L., Till R., Lambert C., Capeness M., Lerner T., Fenton A., Barrow P., and Sockett E. “Effects of Orally Administered Bdellovibrio bacteriovorus on the Well-being and Salmonella Colonization of Young Chicks.” Applied and Environmental Microbiology, 2011, DOI: 10.1128/AEM.00426-11
11Coburn B., Sekirov I., and Finlay B. “Type III Secretion Systems and Disease.” Clinical Microbiology Reviews, 2007, DOI: 10.1128/CMR.00013-07
Bacteria Genomes BDELLOVIBRIO BACTERIOVORUS. (2006, August 30). 2Can Support Portal: Genomes- All Genomes. Retrieved November 3, 2006, from http://www.ebi.ac.uk/2can/genomes/genomes.html?http://www.ebi.ac.uk/2can/genomes/bacteria/Bdellovibrio_bacteriovorus.html
Bacterial Nomenclature Up-to-Date. (2004). DSMZ- the German Resource Centre for Biological Material. Retrieved November 3, 2006, from http://www.dsmz.de/microorganisms/bacterial_nomenclature_info.php?genus=Bdellovibrio&show_all_details=1
Bdellovibrio bacteriovorus. (2006). MEROPS- the peptidase database. Retrieved November 3, 2006, from MEROPS database: http://merops.sanger.ac.uk/cgi-bin/speccards?sp=sp002161&type=P
Bdellovibrio genome sequenced. (2006). Genome Biology Research News. Retrieved November 3, 2006, from http://genomebiology.com/researchnews/ default.asp?arx_id=gb-spotlight-20040130-01
Fox, J. F. (2006). Bdellovibrio make fast food of gram-negative bacteria . In Microbe Magazine. Retrieved December 10, 2006, from http://www.asm.org/ microbe/index.asp?bid=44300
Lambert, C., Smith, M. C. M., & Sockett, R. E. (2003, February). A novel assay to monitor predator-prety interaction for Bdellovibrio bacteriovorus 109 J reveals a role for methylaccepting chemotaxis proteins in predation. Environmental Biology, 5(2), 127. Abstract retrieved November 3, 2006, from http://www.blackwell-synergy.com/links/doi/10.1046/ j.1462-2920.2003.00385.x
Shemesh, Y., Davidov, Y., Koval, S., & Edouard, J. (2003). [Review of the scientific experiment Small eats big: ecology and diversity of Bdellovibrio and like organisms, and their dynamics in predator-prey interactions]. EDP Sciences, 23, 433-439. Retrieved November 3, 2006, from http://www.edpsciences.org/articles/agro/pdf/2003/05/A3505.pdf?access=ok
Schwudke, D., Linscheid, M., Strauch, E., Appel, B., Zahringer, U., Moll, H., et al. (2003, July 25). The Obligate Predatory Bdellovibrio bacteriovorus Possesses a Neurtal Lid A Containing {-D-Mannoses That Replace Phosphate Residues. Biol. Chem, 278(30), 27502-27512. Retrieved November 10, 2006, from jbc ONLINE database: http://www.jbc.org/cgi/content/full/278/30/27502