Cupriavidus metallidurans
Classification
Higher order taxa
Eubacteria (kingdom); Bacteria (domain); Proteobacteria (phylum); Betaproteobacteria (class); Burkholderiales (order); Burkholderiaceae (family); Cupriavidus (genus)
Species
Cupriavidus metallidurans [1]
Description and Significance
Cupriavidus metallidurans (previously Ralstonia metallidurans [2]) type strain CH34 (hereafter C. metallidurans) is a Gram-negative , motile, non-spore forming, rod-shaped bacterium known for its ability to resist toxic heavy metals; metallidurans literally translates to "metal-enduring"[3] [4]. When cultured on trypticase soy agar (TSA) at 30°C for 24h, C. metallidurans forms round, smooth, flat and convex colonies that are transparent and about 0.5mm in diameter. C. metallidurans is a mesophile; it has not been shown to grow under conditions outside of a range of 20-37°C.
This bacterium’s high number of heavy metal resistance genes and ability to thrive in toxic environments make it an important model organism in studying the mechanisms by which diverse microbes deal with heavy metal stress [5]. C. metallidurans is also able to precipitate solid gold from aqueous gold (III) tetrachloride, which is a potent heavy metal toxin to most organisms [4].
Genome Structure
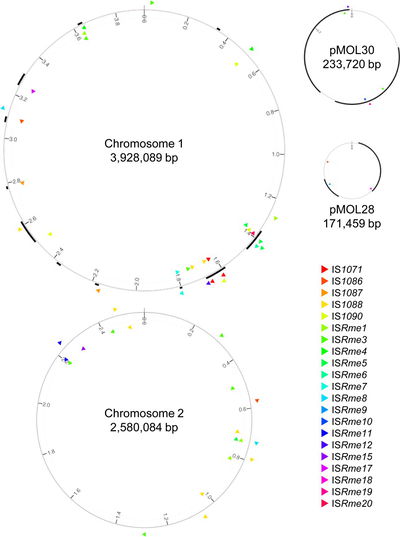
The genome of C. metallidurans comprises two circular chromosomes (known as CHR1 and CHR2) as well as two circular megaplasmids (pMOL28 and pMOL30) [7]. CHR1 (3,928,089 bp, 63.82% G+C) holds the majority of housekeeping genes between the two chromosomes, and thus it is believed to be the original, ancestral replicon. CHR2 (2,580,084 bp, 63.60% G+C), however, was probably acquired as a plasmid, and throughout the evolution of C. metallidurans it was transformed into a second chromosome via gene transfers (either directly from CHR1 or from another source) [1].
Of the two megaplasmids, pMOL28 (171,459 bp, 60.50% G+C) has one genomic island containing operons for metal resistance and contains genes involved in resistance to Co(II), Cr(VI), Hg(II), and Ni(II). pMOL30 (233,720 bp, 60.13% G+C) has two genomic islands and contains genes involved in resistance to Ag(I), Cd(II), Co(II), Cu(II), Hg(II), Pb(II), and Zn(II). Both plasmids contain genes that are both up-regulated and/or down-regulated when the cell is exposed to certain metal cations [7].
Metabolism
C. metallidurans is a facultative anaerobe that can undergo regular oxidative phosphorylation, but when there is limited access to oxygen in the environment, C. metallidurans is also able to reduce nitrate, but not nitrite [3]. C. metallidurans is unable to grow on glucose, fructose, or galactose because it lacks sufficient glucose uptake systems. It can, however, produce energy by assimilating some acidic sugar-derivatives which, via a chain of pathways, will end up producing two molecules of pyruvate each to feed in to the TCA cycle or other related cycles [1]. When organic carbon sources are unattainable, C. metallidurans can become autotrophic by fixing and reducing CO2 to synthesize organic molecules, and has the capacity to oxidize inorganic sulfur compounds as well as hydrogen gas to obtain electrons [1]. The myriad of adaptive genes within the C. metallidurans genome also code for proteins, such as alcohol and aldehyde dehydrogenases, that allow this bacterium to obtain both its carbon and energy from recalcitrant organic compounds, such as propanol, butanol, and acetone when other sources are unavailable [1].
Ecology and Pathology
C. metallidurans is normally found in industrial sediments or wastes which contain high heavy metal concentrations [9]. C. metallidurans was initially isolated from the wastewater of a zinc factory in Liège, Belgium [3]. Later, this bacterium was found in another unusual place; it was the dominant organism present in biofilms on gold grains found in the Prophet gold mine in Queensland, Australia [8]. This bacterium is able to survive in these environments due to the abundance of metal-resistance genes dispersed throughout its genome which code for heavy metal exporters, regulators,and potential metal-binding proteins used to detoxify the cell [10]. These abilities have earned C. metallidurans some fame. When placed in an environment containing aqueous Gold (III) chloride, this bacterium reduces the complex – by a mechanism yet to be fully understood – leading to the precipitation of solid gold nanoparticles within or outside the cell [10].
Until recently, C. metallidurans was thought to be non-pathogenic, but in 2011 a patient with multiple complications after an invasive surgery was shown to have contracted a nosocomial infection with C. metallidurans; the bacterium was repeatedly cultured from the patient’s blood [13].
Two strains of C. metallidurans were also identified from the respiratory secretions of cystic fibrosis patients. The actual pathogenic effects of C. metallidurans are not understood in either of these cases [6].
Applications to Biotechnology
The metabolic and heavy-metal resistant capabilities of C. metallidurans make it an excellent candidate for many biotechnological applications in which nutrient sources are limited and conditions are unfavorable for most organisms.
While the ability of C. metallidurans to precipitate pure gold is a fascinating one, it is not economical on a large scale; considering one requires gold to make gold (III) chloride, there is no real benefit.
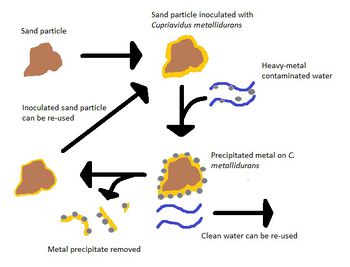
It does, however, have potential in many other areas due to these abilities. One of these areas is the decontamination of heavy metal-containing waste water. By using a sand filter inoculated with C. metallidurans, one can remove these contaminants from the water so it can be recycled [11]. First, sand particles are inoculated with the bacteria and put in contact with the contaminated waste water. The bacteria bound to the sand particle precipitate the dissolved heavy metals into solid form on their membrane or within the cytoplasm. The metal-laden biomass is then removed and the water is free of contaminants [11].
Another application of C. metallidurans is in biosensing[12]. Biosensors are devices that use biological systems to detect specific compounds; the binding of the sensor, which is usually an enzyme, results in a measurable change. Due to the specificity of multiple gene products that are up-regulated in C. metallidurans when exposed to gold, this bacterium is a promising candidate for use as a biosensor. One could theoretically utilize these gold-specific proteins to immediately detect and quantify gold concentrations in environmental samples. This would save much time and money, and be of significant benefit in the gold mining industry [12].
References
1. Janssen P.J., Van Houdt R., Moors H., Monsieurs P., Morin N., Michaux A., Benotmane M.A., Leys N., Vallaeys T., Lapidus A., Monchy S., Médigue C., Taghavi S., McCorkle S., Dunn J., van der Lelie D. and Mergeay M. “The Complete Genome Sequence of Cupriavidus metallidurans Strain CH34, a Master Survivalist in Harsh Anthropogenic Environments.” PLoS ONE, 2010, DOI: 10.1371/journal.pone.0010433
2. Vandamme P. and Coenye T. “Taxonomy of the genus Cupriavidus: a tale of lost and found.” International Journal of Systematic and Evolutionary Microbiology, 2004, DOI: 10.1099/ijs.0.63247-0
3. Goris J., De Vos P., Coenye T., Hoste B., Janssens D., Brim H., Diels L., Mergeay M., Kersters K. and Vandamme P. “Classification of metal-resistant bacteria from industrial as Ralstonia campinensis sp. Nov., Ralstonia metallidurans sp. Nov. and Ralstonia basilensis. Steinle et al. 1998 emend.” International Journal of Systematic and Evolutionary Microbiology, 2001, DOI: 10.1099/00207713-51-5-1773
4. Reith F., Rogers S.L., McPhail D.C. and Webb D. “Biomineralization of Gold: Biofilms on Bacterioform Gold.” Science, 2006, DOI: 10.1126/science.1125878
5. Monsieurs P., Moors H., Van Houdt R., Janssen P.J., Janssen A., Coninx I., Mergeay M. and Leys N. “Heavy metal resistance in Cupriavidus metallidurans CH34 is governed by an intricate transcriptional network.” BioMetals, 2011, DOI: 10.1007/210534-011-9473-y
6. Coenye T., Spilker T., Reik R., Vandamme P. and LiPuma J.L. “Use of PCR Analyses To Define the Distribution of Ralstonia Species Recovered from Patients with Cystic Fibrosis.” Journal of Clinical Microbiology, 2005, DOI: 10.1128/JCM.43.7.3463-3466.2005
7. Monchy S., Benotmane M.A., Janssen P., Vallaeys T., Taghavi S., van der Lelie D. and Mergeay M. “Plasmids pMOL28 and pMOL30 of Cupriavidus metallidurans Are Specialized in the Maximal Viable Response to Heavy Metals.” Journal of Bacteriology, 2007, DOI: 10.1128/JB.00375-07
8. Reith F., Fairbrother L., Nolze G., Wilhelmi O., Clode P.L., Gregg A., Parsons J.E., Wakelin S.A., Pring A., Hough R., Southam G. and Brugger J. “Nanoparticle factories: Biofilms hold the key to gold dispersion and nugget formation.” Geology, 2010, DOI: 10.1130/G31052.1
9. Mergeay M., Monchy S., Vallaeys T., Auquier V., Benotmane A., Bertin P., Taghavi S., Dunn J., van der Lelie D. and Wattiez R. “Ralstonia metallidurans, a bacterium specifically adapted to toxic metals: towards a catalogue of metal-responsive genes.” FEMS Microbiology Reviews, 2003, DOI: 10.1016/SO168-6445(03)00045-7
10. Reith F., Etschmann B., Grosse C., Moors H., Benotmane M.D., Monsieurs P., Grass G., Doonan C., Vogt S., Lai B., Martinez-Criado G., George G.N., Nies D.H., Mergeay M., Pring A., Southam G. and Brugger J. “Mechanisms of gold biomineralization in the bacterium Cupriavidus metallidurans.” Proceedings of the National Academy of Sciences of the United States of America, 2009, DOI: 10.1073/pnas.0904583106
11. Diels L., Van Roy S., Taghavi S. and Van Houdt R. “From industrial sites to environmental applications with Cupriavidus metallidurans.” Antonie van Leeuwenhoek, 2009, DOI: 10.1007/s10482-009-9361-4
12. Zammit C.M., Cook N., Brugger J., Ciobanu C.L. and Reith F. “The future of biotechnology for gold exploration and processing.” Minerals Engineering, 2012, DOI: 10.1016/j.mineng.2012.03.016
13. Langevin S., Vincelette J., Bekal S. and Gaudreau C. “First Case of Invasive Human Infection Caused by Cupriavidus metallidurans.” Journal of Clinical Microbiology, 2011, DOI: 10.1128/JCM.01947-10