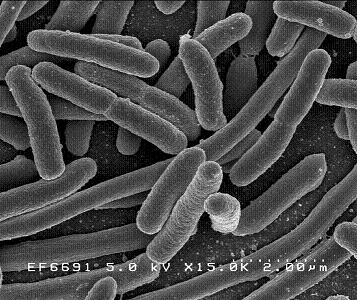
Escherichia coli
A Microbial Biorealm page on the genus Escherichia coli
Classification
Higher order taxa
Eubacteria (Kingdom); Bacteria (Domain); Proteobacteria (Phylum); Gamma Proteobacteria (Class); Enterobacteriales (Order); Enterobacteriaceae (Family); Escherichia (Genus)
Species
Escherichia coli
Description and Significance
E. coli was first discovered in 1885 by Theodor Escherich, a German bacteriologist. E. coli has since been commonly used for biological lab experiment and research. E. coli is a facultative (aerobic and anaerobic growth) gram-negative, rod shaped bacteria that can be commonly found in animal feces, lower intestines of mammals, and even on the edge of hot springs. They grow best at 37 C. E. coli is a Gram-negative organism that can not sporulate. Therefore, it is easy to eradicate by simple boiling or basic sterilization. E. coli can also be classified into hundreds of strains on the basis of different serotypes. E. coli O157:H7, for example, is a well-studied strain of the bacterium E. coli, which produces Shiga-like toxins, causing severe illness by eating cheese and contaminated meat[1]. Furthermore, enteric E. coli can be classified into six categories based on its virulence properties, such as enterotoxigenic E. coli (ETEC), enteropathogenic E. coli (EPEC), enteroinvasive E. coli (EIEC), enterohemorrageic E. coli (EHEC), enteroadherent aggregative E. coli (EAggEC), and verotoxigenic E. coli (VTEC)[2]. These enteric E. coli can cause several intestinal and extra-intestinal infections such as urinary tract infection and mastitis. However, E. coli are not always harmful to human bodies or other animals. Most E. coli live in our intestines, where they help our body breakdown the food we eat as well as assist with waste processing, vitamin K production, and food absorption.
Genome structure
E. coli has only one circular chromosome, some along with a circular plasmid. Its chromosomal DNA has been completely sequenced by lab researchers. E. coli has a single chromosome with about 4,600 kb, about 4,300 potential coding sequences, and only about 1,800 known E. coli proteins. 70% of the chromosome is composed of single genes (monocistronic), and 6% is polycistronic. About 30% of the sequenced ORF's (Open Reading Frames, areas that look like they could be protein coding genes) have unknown functions[3].
Moreover, there are many different strains of E. coli; each of these strains differs in its genotype from wild-type E. coli. The genotype will then affect the phenotype that is expressed, and further influences the physiology and life cycle of each strain. Therefore, different strains of E. coli can live in different kinds of animals. The natural biological process of mutation in genomes is the major cause to produce so many different strains of E. coli. In addition, similar to most bacteria, E. coli can transfer its DNA materials through bacterial conjugation with other related bacteria to produce more mutation and add more strains into the existing population.
Cell Structure and Metabolism
E. coli is a Gram-negative rod-shaped bacteria, which possesses adhesive fimbriae and a cell wall that consists of an outer membrane containing lipopolysaccharides, a periplasmic space with a peptidoglycan layer, and an inner, cytoplasmic membrane. Some strains are piliated and capable of accepting and transferring plasmid to and from other bacteria. Such property enables E. coli under bad/stress conditions to survive. [4] Even though it has extremely simple cell structure, with only one chromosomal DNA and a plasmid, it can perform complicated metabolism to maintain its cell growth and cell division.
Cell structure of E. coli from Wikipedia.[5]
E. coli can produce soluble amyloid oglimers and precipitate them as curli because E. coli have a specific nucleation-precipitation machinery. The network of fibers can bind E. coli to their host cells. However, the significance of amyloid in E. coli is still unknown. [6]
Ecology
Escherichia coli can be commonly found in lower intestines of human and mammals. When E. coli locates in human large intestines, it can help digestion processes, food breakdown and absorption, and vitamin K production. Different strains of E. coli can be found in different type of animals, so we can determine the source (from human or from other animals) of the stools by examining which strain of E. coli is present in the stools. E. coli can also be found in environments at higher temperature, such as on the edge of hot springs.
E. coli is commonly used as an indicator in the field of water purification. The E. coli-index can indicate how much human feces is in the water. The reason why E. coli is used as an indicator is due to a significant larger amount of E. coli in human feces than other bacterial organisms. Most strains of "E. coli" are not harmful to their hosts; however, more and more newly discovered strains are contributing into existing population through mutation and evolution. Some can cause severe disease, such as E. coli O157:H7.
Pathology
Although E. coli in human large intestine can assist with waste processing and food absorption, some strains of E. coli can cause severe infections in many animals, such as humans, sheep, horses, dogs, etc. The one that only found in humans is called enteroaggregative E. coli. Urinary tract infection, for example, can be caused by ascending infections of urethra. Such infections can be found in both adult male and female, and some infants can be infected as well.
E. coli O157:H7 is one of the most infective strains that can cause food poisoning. It belongs to enterohemorrhagic strain of the E. coli and can lead to bloody diarrhea and kidney failure when one gets infected by contaminated ground beef, unpasteurized milk or contaminated water. The toxin that E. coli O157:H7 produces is a Shiga-like toxin which is a regulated toxin that catalytically inactivate 60S ribosomal subunits of most eukaryotic cells, blocking mRNA translation and thus causing cell death.[7] Some important symptoms are diarrhea that is acute and severe, either bloody or not bloody, stomach cramping, vomiting, loss of appetite, abdominal pain, and fever. The causes can usually clear up on their own in 1-3 days with no treatment required. However, patients should avoid dairy products because those products may induce temporary lactose intolerance, and therefore make the diarrhea worse.
Application to Biotechnology
E. coli plays an important role in current biological engineering because of its manipulation and long laboratory history. It has been widely used to synthesize DNA and proteins. Most results from E. coli research can be applied to animals and humans. The most useful contribution of recombinant DNA from E. coli is to use the manipulation of E. coli to produce human insulin for diabetes patients. Since E. coli can produce human enzymes through recombinant DNA techniques, it is widely used as a very good tool to produce useful compounds or enzymes for medication. In a recent study (Apr. 2007), scientists has explored a new method to cure Alzheimer's disease (AD), which is a neurodegenerative disorder characterized by a progressive loss of cognitive function due to extra deposition of the longer form of the amyloid peptide Abeta. In this study, scientists use E. coli to enable rapid production of the antigen and its purification.[8] In other words, E. coli plays an important role to enable the rapid, continuous production and purification in large amount of human Abeta sequence by its unique expression system.
Current Research
One recent study is working on the adherent-invasive E. coli (AIEC) which can abnormally colonize the ileal mucosa of Crohn disease (CD) patients and adhere to and invade intestinal epithelial cells of CD patients. The study shows that this kind of CD-associated AIEC strain can adhere to the brush border of primary ileal enterocytes isolated from CD patients. The adhesion of AIEC is dependent on type 1 pili expression on baterial surface and on carcinoembryonic antigen-related cell adhesion molecule 6 (CEACAM6) expression on the apical surface of ileal epithelial cells. CEACAM6 is an essential receptor for AIEC strain to adhere the ileal eithelial cells in CD patients. Moreover, this study also performs in vitro experiments indicating that AIEC can boost its own colonization in CD patients.[9]
Another recent study is focusing on exploring the antigens on the outer membrane of the uropathogenic E. coli (UPEC) which can cause uncomplicated urinary tract infection (UTI), and furthermore to design a UTI vaccine to promote protective immunity against UPEC infection. In this sutdy, they apply an immunoproteomics approach to vaccine development that has been used successfully to identify vaccine targets in other pathogenic bacteria. The outer membrane proteins of UPEC from infected mice are separated by two-dimensional gel electrophoresis and are identified by mass spectrometry. A total of 23 antigens have known roles in UPEC pathogenesis, such as ChuA, IroN, IreA, Iha, IutA, and FliC. After identifying the antigens on outer membrane of UPEC, they demonstrate an antibody targeting directly on these antigens during UTI. This study shows that these conserved outer membrane antigens can be used as rational candidates for a UTI vaccine. [10]
Another recent study is focusing on genomic evolution of E. coli O157:H7 strains which diverge into two distinct lineages, lineages I and lineages II and appear to present different ecological characteristics. Lineage I strains are more commonly associated with human disease than lineage II. This experiment is carried out by microarray-based comparative genomic hybridization (CGH) to identify genomic differences among 31 E. coli O157:H7 strains which have different lineage-specific polymorphism assay types. Among 31 E. coli O157:H7 strains, there are 15 lineage I, 4 lineageI/II, and 12 lineage Ii strains, respectively. From the CGH data, they conclude that the presence of two dominant lineages subgroup E. coli O157:H7. The genomic composition of these subgroups suggests that the genomic divergence and lateral gene transfer have contributed to the evolution of E. coli. In addition, the genomic differences between lineage I and lineage II may contribute to extinct epidemiology and ecology of different strains of E. coli O157:H7. [11]
Another study involving E. coli involves the uniqueness of gram negative bacteria in the fact that they have two membranes for materials to cross in order to get in or out of the cell. The inner membrane and the outer membrane provide a barrier that is extremely proficient at maintaining homeostasis. Although the inner membrane is self-energized by proton motive force across it, the outer membrane is not self-energized, and thus must obtain energy to import and export essential substances, such as iron siderophores. Two protein systems in E. coli are known to transfer energy from the inner membrane to the outer membrane: the Ton and Tol systems. The Ton system, consisting of TonB, ExbB, and ExbD is better studied than the Tol system, consisting of TolA, TolQ, and TolR. These systems both consist of an energy harvesting complex embedded in the inner membrane of E. coli and a protein that the energy gets transferred to which in turn energizes the outer membrane. In the Ton system, ExbB and ExbD heteromultimers make up the energy harvesting complex that transfers energy to TonB, which undergoes a conformational change and provides energy for a variety of functions in the outer membrane. Similarly in the Tol system, TolQ and TolR are embedded in the inner membrane as the energy harvesting complex in heteromultimers and transfer energy to TolA, which undergoes a conformational change in order to supply energy to the outer membrane. While the Ton system has been shown to be involved with transport of materials across the membrane, the Tol system has been shown to be involved in maintenance of the outer membrane. Continued research involving these two systems could play a role in developing new antibiotics that target gram negative bacteria and their ability to facilitate the uptake of essential nutrients, as well as the extrusion of harmful substances. [13, 14, 15, 16]
A long-term study has been done on genome evolution and adaptation of Escherichia coli. Comparative genome sequencing done on an experimental population of E. coli has allowed for further investigation between the relationship between genome evolution and organismal adaptation. E. coli was grown and sampled for almost 20 years with genome sequencing at generations 2,000, 5,000, 10,000, 15,000, 20,000 and 40,000 from an asexual population that evolved with glucose as a limiting nutrient. Comparative genome sequencing showed mutational differences between the ancestral and evolved genomes accumulating in a near-linear fashion, whereas the fitness trajectory was strongly nonlinear. In particular, the rate of fitness improvement decelerated over time. The discordance in rates of genomic and adaptive evolution cannot be explained by the drift hypothesis, but rather the discrepancy may be due to clonal interference, compensatory adaptation, or changing mutation rates. In clonal interference, sub-lineages with the most beneficial mutations outcompete sub-lineages bearing mutations that are not as beneficial. Most beneficial mutations dominate the early phase of evolution for large populations in a new environment, but there are more potential mutations that confer small advantages than large (adaptative) ones. The population in the study retained a low ancestral mutation rate to at least 20,000 generations but in later generations, however, this population exhibited a greatly elevated rate of genomic evolution when a mutator lineage became established later. Furthermore, there were no synonymous mutations fixed in the first 20,000 generations and this is consistent with the low point-mutation rate in E.coli and population-genetic theory. These results indicate that it is important to explore long-term dynamic coupling between genome evolution and adaptation. [17]
References
1. Atlanta, Ga. "Foodborne Disease Outbreak Investigation, Epidemiologic Case Study." p. 9 -17. <http://roger.ucsd.edu:80/record=b4800216 > Retrieved on 2007-5-10.
2. Parry, Sharon. " E. coli: Environmental Health Issues of VTEC O157." London; New York: Spon Press, 2002 <http://www.engnetbase.com/books/2452/tf5952_fm.pdf> Retrieved on 2007-5-11.
3. Reed, Jennifer Leanne. "Model Driven Analysis of Escherichia coli Metabolism." 2005. <http://roger.ucsd.edu/search/Xe.+coli%2C+genome&searchscope=7&SORT=D/Xe.+coli%252C+genome&searchscope=7&SORT=D&extended=1&SUBKEY=e.%20coli%252C%20genome/1%2C3%2C3%2CE/frameset&FF=Xe.+coli%252C+genome&searchscope=7&SORT=D&2%2C2%2C> Retrieved on 200-5-18.
4. National BioResource Project.<http://www.shigen.nig.ac.jp/ecoli/strain/top/top.jsp> Retrieved on 2007-5-28.
5. Wikipedia. <http://en.wikipedia.org/wiki/Image:EscherichiaColi_NIAID.jpg> Retrieved on 2007-5-28.
6. Medical Encyclopedia. <http://www.nlm.nih.gov/medlineplus/ency/article/000296.htm> Retrieved on 2007-5-20.
7. Champman MR, Robinson LS. "Role of E. coli Curli Operons in Directing Amyloid Fiber Formation." Feb. 2002. <http://www.ncbi.nlm.nih.gov/sites/entrez?cmd=Retrieve&db=pubmed&dopt=Abstract&list_uids=11823641> Retrieved on 2007-6-2.
8. Philpott, Dana. "E. coli: Shiga toxin methods and protocols." <http://roger.ucsd.edu:80/record=b4427335> Retrieved on 2007-5-19.
9. Subramanian S, Shree AN. "Expression, Purification and Characterization of a Synthetic Gene Encoding Human Amyloid Beta in Escherichia coli.<http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=pubmed&cmd=Retrieve&dopt=AbstractPlus&list_uids=17536333&query_hl=14&itool=pubmed_docsum> Apr. 2007. Retrieved on 2007-5-25.
10. Barnich N, Carvalho FA, Glasser AL, Darcha C, Jantscheff P, Allez M, Peeters H, Bommelaer G, Desreumaux P, Colombel JF, Darfeuille-Michaud A. "CEACAM6 Acts as a REceptor for Adherent-invasive E. coli, Supporting Ileal Mucosa Colonization in Crohn Disease. May 24, 2007. <http://www.ncbi.nlm.nih.gov/sites/entrez?Db=pubmed&Cmd=ShowDetailView&TermToSearch=17525800&ordinalpos=7&itool=EntrezSystem2.PEntrez.Pubmed.Pubmed_ResultsPanel.Pubmed_RVDocSum> Retrieved on 2007-6-1.
11. Hagan EC, Mobley HL. "Uropathogenic E. coli Outer Membrane Antigens Expressed During Urinary Tract Infection." May 21, 2007. <http://www.ncbi.nlm.nih.gov/sites/entrez?Db=pubmed&Cmd=ShowDetailView&TermToSearch=17517861&ordinalpos=10&itool=EntrezSystem2.PEntrez.Pubmed.Pubmed_ResultsPanel.Pubmed_RVDocSum> Retrieved on 2007-6-1.
12. Zhang Y, Laing C, Steele M, Ziebell K, Johnson R, Benson AK, Taboada E, Gannon VP. "Genome Evolution in Major Escherichia coli O157:H7." May 16, 2007. <http://www.ncbi.nlm.nih.gov/sites/entrez?Db=pubmed&Cmd=ShowDetailView&TermToSearch=17506902&ordinalpos=16&itool=EntrezSystem2.PEntrez.Pubmed.Pubmed_ResultsPanel.Pubmed_RVDocSum> Retrieved on 2007-6-1.
13. Brinkman, Kerry K. and R.A. Larsen. 2008. Interactions of the energy transducer TonB with noncognate energy-harvesting complexes. Journal of Bacteriology. 190 (1): 421-427.
14. Keller, Kimberly L, K.K. Brinkman, and R.A. Larsen. TonB/TolA amino-terminal domain modeling. 2007. Methods in Enzymology. 23005 (5): 134-148.
15. Larsen, Ray A. G.E. Deckert, K.A. Kastead, S. Devanathan, K.L. Keller, and K. Postle. His20 provides the sole functionally significant side chain in the essential TonB transmembrane domain. (2007). Journal of bacteriology. 189 (7): 2825-2833.
16. Postle, Kathleen and R.A. Larsen. 2007. TonB-dependant energy transduction between outer and cytoplasmic membranes. Biometals. 20:453-465.
17. Barrick, J., et al. “Genome evolution and adaptation in a long-term experiment with Escherichia coli” Nature, 2009. Volume 461. p. 1243-1247.
European Journal of Biochemistry. By Federation of European Biochemical Societies. 2004
Wikipedia <http://en.wikipedia.org/wiki/Escherichia_coli#Strains>
Edited by Nicole Jacques and Nancy Ngo, students of M. Glogowski at Loyola University Chicago
.