Marinobacter aquaeolei
A Microbial Biorealm page on the genus Marinobacter aquaeolei
Classification
Higher order taxa
Bacteria; Proteobacteria; Gammaproteobacteria; Alteromonadales; Alteromonadaceae; Marinobacter; Marinobacter aquaeolei
Species
Marinobacter aquaeolei
|
NCBI: Taxonomy |
Description and significance
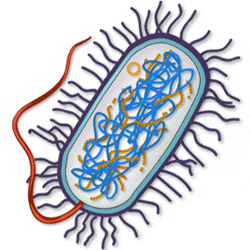
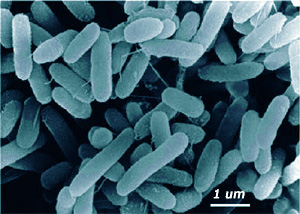
Marinobacter aquaeolei are Gram-negative, non-spore forming, and rod shaped cells. They are motile by the use of a polar flagellum. Several strains of this cell were found at the head of and oil-producing well on an offshore platform in southern Vietnam. Marinobacter aquaeolei can be found throughout the water column and in the deep ocean. These organisms are capable of extremophilic lifestyles and are moderately halophilic and mesophilic. The growth of cells occurred at NaCl concentrations between 0% - 20%; the optimal level is at 5% NaCl.[2]
Iron is one of the most important redox-active elements in the solar system. Iron oxidation is distributed widely among prokaryotes and the activities of these iron oxidizing bacteria is critical to many major elemental cycles, including the carbon cycle. Sequencing the genome of this bacterium can help us understand the fundamental biology of iron oxidation. Marinobacter aquaeolei grows rapidly on plates and in liquid culture and is open to biochemical methods of genetic manipulation.
Genome structure
Marinobacter aquaeolei consists of 3960 genes comprised from 4,326,849 nucleotides. The nucleotides make up one double-stranded circular DNA. The GC content of the bacteria is 57%. The bacteria's chromosomal topology is circular. It also consists of two plasmids, pMAQU01 and pMAQU02[1]. There are also 63 structural RNAs. Like most other microbial organisms, Marinobacter aquaeolei lacks a true nucleus that stores DNA and also transcription. The cell does have a nucleoid which is where the genome is packaged. Transcription takes place between the cytoplasm and nucleoid. Because Marinobacter aquaeolei is known as an oxidator of iron, its DNA contains specific nucleotide sequences that code for genes that produce the system of enzymes that are necessary for iron oxidation.
A 16S rDNA sequence analysis showed that Marinobacter aquaeolei and Marinobacter hydrocarbonoclasticus were very closely related with a 99.4 % similarity.[2] Even isoprenoid quinones, antibiotic resistances, and fatty acid compositions were similar. The difference was found in the whole-cell protein pattern. Further deviations could be found in Fourier transformed IR spectra of cells and LPS composition. [2]
Cell structure and metabolism
Rod shaped isolates were obtained from offshore oil wells and all were Gram-negative cells. They were able to grow in a wide range of NaCl mediums. They were found to grow singly or in pairs. The cells were actively mobile by a polar flagellum only visible in some areas. "dl-Lactate, acetate, succinate, fumarate, citrate, l-glutamate and butyrate could be used as sole carbon and energy sources..." (Huu NB, Denner EB, Ha DT, Wanner G, Stan-Lotter H. Marinobacter aquaeolei can also use pristane, n-hexadecane, and crude oil components as sole carbon sources.[2]
As an oxidizer of iron, the kinetics of ferrous iron are known to be pH dependent. Marinobacter aquaeolei must also actively compete with other microbes for available ferrous iron. The organism produces an unusual class of amphiphilic siderophores, iron transporter molecules with both hydrophilic and hydrophobic parts. And these are the mechanisms thought to be unique in marine bacteria.[4]
Ecology
Marinobacter aquaeolei was originally found an isolated from the head of an oil-producing well on an offshore oil platform. Halophilic bacteria are found in environments such as oil and gas fields and special ecosystems. These environments support these bacteria because of their physio-chemical and geochemical conditions. [2] These organisms can be found throughout the water column and in the deep ocean. These microbes belong to a family of hydrocarbon degrading consortia and are able to live in a variety of conditions. They can be psychrophilic, oligotrophic, or halotolerance. Amphiphilic siderophores are the chief method of iron acquisition in these marine bacteria. Low limits of iron limit primary productivity and also carbon sequestration. [4]
Marinobacter aquaeolei can degrade n-hexadecane, pristane and some crude oil components making it useful for research. [2] Since iron is so abundant, the oxidation of it by this particular bacteria proves to be something useful for research. Scientists still have a poor understanding of this major metabolic pathway. Marinobacter aquaeolei is important to understand for both biological iron oxidation and iron acquisition in the ocean.[4]
Pathology
Marinobacter aquaeolei presents no pathogens and is not pathogenic in any type of hosts [5]
Application to Biotechnology
Marinobacter aquaeolei is used primarily for comparative genomics. It is important since the availability of iron is an important part of the carbon cycle in the ocean. A deficiency in iron is widely believed to limit primary productivity. Being able to study Marinobacter aquaeolei is useful since the fundamental basics of iron oxidation remains one of the least understood of the major metabolic pathways in microbes. Since iron is so abundant in the ocean, studying this particular organism proves to be useful. Understanding this organism fully allows scientists to further understand both the biological oxidation of iron and the acquisition of iron in the ocean. [4]
Iron oxidation, however, happens in our own bodies which is why it is important to understand these iron oxidizers. Iron is an important element in human nutrition. An excess and deficiency in iron both cause problems for the human body. These deficiencies can lead to coronary risk. Available iron in the oxidation sites of humans are questionable. Physiological controls maintain iron in non-reactive forms. Understanding how this bacteria acquires and oxidizes iron can possibly be linked with the iron-heart disease hypothesis. Because of this, a proper diet with iron-rich foods is always recommended.[3]
Current Research
Not much research has been done since completion of the genome for Marinobacter aquaeolei in January of 2007. However, a recent article by Marquez and Ventosa from the Department of Microbiology and Parasitology of the University of Sevilla in Spain, shows how closely related and how different Marinobacter aquaeolei and Marinobacter hydrocarbonoclasticus really are. The research reviews the G+C content, fatty acid composition, and DNA-DNA hybridization studies that were performed to understand the taxonomic positions of these two organisms. Marquez suggests that the two species be united under the same name since they are heterotypic synonyms due to phenotypic and phylogenetic traits. (Marquez, 2005)[1]
Extensive research on Marinobacter aquaeolei is not underway, however, understanding the organism's ability to oxidize iron can lead to understanding of the biological oxidation of iron and also the ability the cell has to acquire iron for use. Iron oxidation is also a fundamental process found in the human body. A study on the importance of iron sufficiency was done by Lyda Associates, regarding iron oxidation and health. The report stated that iron is a necessary element found in the human body and any excess or deficiency of it can lead to bodily malfunctions and even heart disease. Iron is a highly reactive substance that can serve as both an electron donor and an electron acceptor. Important organs such as the kidneys, liver, and bone marrow all need iron to function. (Lyda Associates, 1994) [3]
Marinobacter hydrocarbonoclasticus is observed to synthesize isoprenoid wax ester storage compounds while growing on phytol. Phytol represented the only carbon source and the organism grew under limited nitrogen and/or phosphorous conditions. A recently released draft of Marinobacter aquaeolei was used to search for certain candidate genes, since the two organisms are so closely related. They searched for WS/DGAT and acyl-CoA synthetase genes within the genome. It was discovered that one of the Marinobacter hydrocarbonoclasticus wax ester synthases exhibited "several orders of magnitude higher activity toward acyl substrates than any previously characterized acyl-WS..."(Abstract, Holtzapple E, Schmidt-Dannert C.). This showed that they adapted to available carbon sources in the environment.[6]
The structure of the Gram-negative Marinobacter aquaeolei was determined by growing it on a mineral media supplemented with ammonium acetate or hydrocarbons. An unsaturated fatty acid composition meant that there was an aerobic desaturation pathway for acids. An increase of the proportions of saturated fatty acid for Marinobacter aquaeolei would decrease the temperature transition of lipids for maintaining the inner membrane fluidity. [7]
References
[3] Lyda Associates, Inc. : Iron Oxidation and Health, June 1994, Research report 100-5.
[4] Joint Genome Institute Why Sequence Marinobacter aquaeolei?
[5] Marinobacter aquaeolei VT8, complete genome, NCBI.
[8] Marinobacter aquaeolei VT8 Genome Portal Site.
Edited by Allan Ji, student of Rachel Larsen and Kit Pogliano
KMG
