Mycoccus xanthus
Classification
http://cmgm.stanford.edu/~kaiserla/about_myxo/about_myxococcus.html
Higher order taxa
Bacteria: Protobacteria; Delta protobacteria; Myxococcales; Cystobacterineae;, Myxococcaceae; Myxococcus
Species
Soil Bacterium, Delta group of the protobacteria
Description and significance
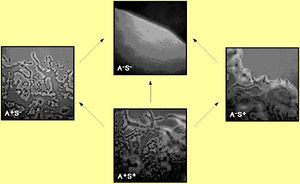
Myxococcus xanthis is a gram negative, rod shaped bacteria. This bacteria moves by gliding motility. There is not much known about this type of motility. There are two different systems which control gliding. The social (S) motility system and the adventurous (A) motility system. The A-cells move more independently. When the cells are close together the S-motility becomes activated (see photo). Myxococcus xanthus was isolated in 1892 by Robert Thaxter. It is capable, during nutrient deprivation, of changing shape. It can go from a vegetative rod to a spherical spore which is a stress resistant spore. The spore is located inside a dome shaped, multicellular fruiting body. When experiencing starvation, the bacterium undergoes a process in which approximately 100,000 cells aggregate to form the fruiting body. This incurs over several hours. The structure becomes spherical spores with thick walls.
Genome structure
Lipolytic enzymes in 3 different families are encoded by the genome of Myxococcus xanthus. The three different families are: patatin lipases, a/B hydrolases, and GDSL lipases. Analysis using gene expression with translational lacZ fusions shows that during development all 3 genes are expressed but MXAN_5522 and MXAN_4569 are the only ones expressed during vegetative growth. By using T7 RNA polymerase transcription (pET 102/D-TOPO) the proteins in these genes were over expressed. The enzymes yielded the highest activity with p-nitrophenyl acetate when by preferentially hydrolyzing esters of short-chain fatty acids.
Cell structure and metabolism
Myxococcus xanthus cohesion involves the interaction of an extra cellular matrix component and a cell surface cohesion. Two defective mutants have been characterized in this work. The fbdA and fbdB are necessary for production cohesions but do not encode them.
Ecology
Myxococcus xanthus is a bacteria found in soil. Decaying vegetation is also a natural habitat. Their microbial cells can be degraded due the fact they are motile by gliding. This forms flat spreading colonies. They are a source of secondary metabolites. These bacteria often exhibit biological activities that are not often found in nature. The bacteria also enters a process of multi cellular development when responding to amino acid starvation. They enter this multicellular association in both feeding and dormant stages. They are the only bacteria to do this. They are also high G&C.
Pathology
Myxococcus xanthus is a soil residing predatory bacterium. During the vegetative part of its life cycle, it moves in colonial swarms. It is thought that the range of microbial prey is enhanced because of the swarm’s collective secretion of biocidal factors. An example is Serratia marcescens. Predation becomes a part of M. xanthus life when certain amino acids and central metabolic intermediates become scarce. Collective group action and mobility are a major part of predation for M. xanthus.
Application to Biotechnology
Approximately equal amounts of polysaccharide and protein make up the cell surface associated structures called fibrils. When cells entered the stationary phase of growth, wild type exopolysaccharides greatly increased. This also took place when Ca2+ was added to growing cells. The polysaccharide production was needed for fruiting-body formation. The exopolysaccharides are also needed to cope with the environment. They are also important in plant and animal bacterial infections.
Current Research
One recent study is identifying major sporulation proteins of Myxococcus xanthus. The proteome of liquid-grown vegetative cells were compared with the proteome of mature fruiting body spores. Protein S and protein S1 were expressed in spores differentially. Three previously uncharacterized proteins were identified. They exhibit no homology to known proteins. The three major proteins were mspA, mspB and mspC. The genes which encoded these spore proteins were inactivated by insertion mutagenesis. The three mutants were able to aggregate. The fruiting bodies of two of the strains remained flattened cell mounds.
Another recent study refers to Cohesion-Defective Mutants. Interaction of a cell surface cohesin with a component fo the extracellular matrix is the result of cohesion of M. xanthus cells. The fbdA and fbdB genes are necessary for their production but do not decode the cohesins. PilA is not considered a major cohesin because both of these nutants produce type IV pili.
One more study refers to the regulation of dev. This is an operon the includes genes that are important for the development of M. xanthus. Spore differentation inside of M. xanthus fruiting bodies is triggered by expression of dev genes. Strong negative autoregulaton occurs in the operon during development. The results of this testing suggest that regulatory elements, either upstream or downstream, interact fucntionally.
References
Journal of Bacteriology, April 2007,Lipolytic Enzymes in Myxoxoxxus xanthus, p. 3072-3080, Vol. 189, No. 8 0021-9193/07/$08.00+0 doi:10.1128/JB.01772-06 http://jb.asm.org.cgi/content/abstract/189/8/3072
Journal of Bacteriology, January 2007,B-D-Allose Inhibits Fruiting Body Formation and Sporulation in Myxococcus xanthus, p. 169-178, Vol. 189. No.1 0021-9193/07/$08.00+0 doi:10.1128/JB.00792-06 http://jb.asm.org.cgi/content/abstract/189/1/169
Journal of Bacteriology, April 2007,Identification of Major Sporulation Proteins of Myxococcus xanthus Using a Proteomic Approach, p.3187-3197, Vol. 189, No. 8 0021-9193/07/$08.00+0 doi:10.1128/JB.01846-06 http://jb.asm.org/cgi/content/abstract/189/8.3187
Journal of Bacteriology, May 2007,Regulation of dev, an Operon That Includes Genes Essential for Myxococcus xanthus Development and CRISPR-Associated Genes and Repeats, p. 3738-3750, Vol. 189, No. 20 0021-9193/07/$08.00+0 doi:10.1128/JB.00187-07 http://jb.asm.org.cgi.content.abstract.189/10/3738
Mutlicellular development in Myxobateria, “Gene Regulation of A-signal production” 11/12/2007 http://www.ucs.mun.ca/~barretta/homepage.html
The Myxobacteria, “What are the Myxobacteria?” November 12, 2007 http://myxobacteria.ahc.umn.edu.whataremyxos1.html
About Myxococcus xanthus, November 12, 2007-11-18 http://cmgm.stanford.edu/~kaiserla/about_myxo/about_myxococcus.html
The unique DKxanthene secondary metabolite family from the myxobaterium Myxococcus xanthus is required for developmental sporulation, November 12, 2007 http://www.pnas.org/cgi/content/abstract/103/50/19128
Mutations affecting predation ability of the soil bacterium Myxococcus xanthus http://mic.sgmjournals.org/cgi/reprint/151/6/1865.pdf
TIGR CMR Comprehensive Microbial Resource Myxococcus xanthus DK1622 Genome Page http://cmr.tigr.org/tigr-scripts/CMR/GenomePage.cgi?org=gmx
http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=93539
Edited by student of Rachel Larsen