Prion Propagation
by Ken Farabaugh
Prions are unique infectious agents that consist only of a protein, and are not replicated via nucleic acid. The wild type protein PrPC (Prion-related Protein, shown in Figure 1 A below) consists of approximately 253 amino acids (split evenly between an N-terminal domain and a C-terminal domain) in primarily alpha-helix secondary structures with one disulfide bond (between Cys179 and Cys214).[1] The prion protein is expressed throughout the body, including fibroblasts and keratinocytes in the skin,[2] though at highest levels in the central nervous system.[3] The function is unclear, although knockout mice strains indicate that it is necessary for survival of the organism. Some scientists theorize that PrPC is involved in copper uptake or ROS tolerance.[3] PrPC is a cell surface protein held in place by a glycolipid anchor attached to Asn231.[1] However, due to either a mutation in the PRNP gene sequence, the amino acid sequence, or error in posttranslational protein folding, the PrPC protein is misfolded into an isoform PrPSc (shown in Figure 1 B below), which consists of mainly beta-sheet fibrillar secondary structures that tend to aggregate, also known as a form of amyloid. This alteration in secondary structure leads to accumulation of PrPSc, formation of plaques, and prion diseases such as Creutzfeldt-Jakob disease (CJD), Gerstmann-Straussler-Scheinker disease, kuru, Bovine Spongiform Encephalopathy (BSE or "mad cow" disease), scrapie, and Familial Fatal Insomnia (FFI). Since the aggregates are formed from native protein sequences, no immune response is raised, and since the plaques assume a non-standard conformation, they are semi-resistant to proteinase K (PK) degradation.[4] The normal half-life of a PrPC is approximately 3-6 hours, but proteinase K resistance increases this time.[5] Polysaccharides have also been shown to play a role in PrPSc structure, and sphingolipids and cholesterol have even been found in purified preparations of prion rods in scrapie (a prion disease found in sheep).[4]

History of the Prion
Scrapie was first discovered in sheep in 1732. Later, amyloid plaques were described in 1854 by Prussian pathologist Rudolph Virchow as 'starch-like', but were proven to consist of proteinaceous material. It wasn't until 1920 that Hans Gerhard Creutzfeldt described a 23-year old woman with a fatal neurological disease, and in 1921 Alfons Maria Jakob described three more patients with the same disease, characterized by neuronal degeneration and gliosis (proliferation of glial cells, non-neuronal cells in the CNS that aid in synaptic transmission and signaling transduction pathways)in the cortical, pyramidal and extrapyramidal systems. Creutzfeldt-Jakob disease, as it came to be known, was theorized by Icelandic virologist Bjorn Sigurdsson to be caused by a 'slow virus', characterized by long incubation periods (decades) followed by short clinical periods, and limited to a single tissue in a single host. In 1957, kuru was discovered among the Fore tribe in Papua New Guinea, and in 1959 comparisons were drawn between the infectious natures of scrapie, CJD, and kuru, termed 'subacute spongiform encephalopathies'. In 1966, Daniel Carleton Gajdusek successfully transmitted kuru to a chimpanzee, for which he won the Nobel prize in 1976, and in 1968 CJD was also successfully transmitted to a chimpanzee by Gibbs. Meanwhile, unnoticed, in 1967 Griffith published a paper hypothesizing a proteinaceous infectious agent. It was not until 1982 that Stanley B. Prusiner coined the word 'prion' (proteinaceous infectious particle) to refer to the protein discovered to be associated with scrapie. Prusiner won the Nobel prize in 1997 for his discovery of the prion protein. The prion hypothesis, or protein-only hypothesis, was then set forth to distinguish prions from other forms of nucleic acid-based disease.[5]
Until 1985, prions were an obscure tidbit of knowledge with little real impact in world events. Then a new prion disease was discovered in cattle in Britain, and named Bovine Spongiform Encephalipathy, or "mad cow" disease. Incidences of the disease grew, fear of transmission to humans spread, reaching a height in 1996 when 10 cases of a new variant CJD strain were reported, vCJD. Research increasingly points to the theory that vCJD arose in humans from ingestion of brain matter from BSE-infected cattle. Once the cause was established, incidences decreased again with increased control of cattle slaughtering, rendering, and bovine meal preparation. Although oral intake of prions is 100,000 times less effective in transmission than direct injection to the CNS, over half a million cattle were killed to prevent the spread off the disease. Specified risk material is still found in beef intended for human consumption.[5]
The Prion Hypothesis
The prion hypothesis, which states that prions propagate themselves without nucleic acid involvement, is controversial because it contradicts the fundamental tenet of molecular biology: proteins are translated from RNA which is transcribed from DNA. However, while the precise mechanism of prion propagation is unclear, there is much evidence to support the theory that PrPSc is the primary, if not sole agent responsible for propagation of more PrPSc, not the least of which is a lack of identification of nucleic acid involvement after over two decades of research. Different hypotheses of this interaction exist, most of which involve the PrPSc protein acting as a template to induce misfolding of PrPC, or a mysterious as-yet unidentified “protein X”, which binds one unit of PrPC and one unit of PrPSc together in a complex (shown in Figure 2). Three known classes of prion propagation have been identified as distinct, including sporadic (85% of cases), acquired (5%), and inherited (10%) PrPsc accumulation.[1],[4]
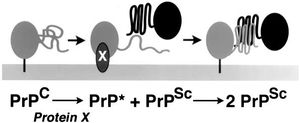
Attempts by researchers to recreate PrPSc using PrPSc from a donor organism of the same species have been well documented, and result in about a 50% successful transmission rate. One of the most supporting pieces of evidence in favor of the protein-only hypothesis is that when mutant PrPSc from a donor organism is inoculated into a recipient organism of the same species, the PrPSc that results has the amino acid sequence of the recipient organism's PrPC, not the donor organism's PrPSc.[4]
Attempts by researchers to recreate PrPSc formation de novo have been conducted, though perhaps raise more questions than they answer. PrPSc formation has been shown to be induced by polyanionic structures such as RNA and polysaccharides in brain homogenates. Attempts to induce PrPSc from mutations in PrPC via an increase in PK-resistant and beta-rich sequences resulted in a number of different isoforms, also indicating that PrPSc formation can depend on intracellular conditions such as solvent concentration and the presence of anionic cofactors.[12] Fibrillar aggregates were induced from PrPC using SDS, certain lipids, and other denaturing agents. However, the initially low infectivity and increasing infectivity corresponding to successive inoculations of these synthetically made prions have led to two hypotheses: the 'maturation' hypothesis, which states that perhaps the amyloid fibrils generated de novo were semi-infectious intermediates between PrPC and PrPSc formation, and the 'adaptation' hypothesis, which states that because the synthetic PrPSc is conformationally different from the PrPC produced in vivo, a 'transmission barrier' develops, which prohibits the propagation of a strain of PrPSc until it adapts enough of the correct conformational modifications of different isoforms of synthetic PrPSc to allow for natural conformational alteration of wild type PrPC. The 'adaptation' model may be supported by evidence that PrPSc aggregates tend to accumulate mutations over their extremely long life-span, resulting from oxidative damage, deamidation, and enzymatic modifications.[4]
Some researchers remain convinced that a bacteria,[14] a virus or virion particle is responsible for all classified prion diseases, despite a lack of evidence in support of this theory. The main problems with this theory include the facts that no virus has yet been identified to be linked with these diseases, features are not consistent with viral encephalitis, and that a percentage of cases do seem to be dominant autosomally inherited in their propagation.[1]
Sporadic Propagation: Creutzfeldt-Jakob disease
CJD is classified as a neuropathological disorder cause by sporadic prion propagation, although there is evidence for a new form, variant-CJD or vCJD, that is acquired. CJD cannot be absolutely diagnosed until a post mortem autopsy, but symptoms may begin as early in life as 28, though median age is 70; spinal taps and EEGs can rule out other forms of treatable dementia. Symptoms include progressively worsening dementia, loss of memory and muscle functions, myoclonus (muscle jerks), and even blindness, leading into a comatose state which is often accompanied by a form of pneumonia. Patients usually die within a single year of onset of symptoms. There is no cure or treatment for the disease as of 2009.[6]
The sporadic mutation of the PRNP gene results in about 1 case of CJD per million individuals per year, consistent with random mutation statistical analysis. No other known infection presents as uniformly throughout all races and tropical regions.[5] This prion disease, caused by rare genetic event or somatic mutation, seems to be focused on amino acid 129. Several different classifications of the PrPSc have been primarily identified by differential cleavage patterns by proteinase K (shown in Figure 3). These types have been additionally separated based on amino acid sequence with respect to a valine or methionine at position 129 in the PrPSc protein, and then by location and formation of plaques in different regions of the brain. Type 1 PrPSc is characterized by homozygous methionine (MM) at codon 129, and synaptic plaque formation primarily in the cerebral cortex and occipital lobe. Type 2 PrPSc can be homozygous methionine, valine (VV), or heterozygous (MV), and causes mainly perivacuolar plaque formation in the basal ganglia and cerebellum in addition to the cerebral cortex. Type 3 PrPSc is characterized by MV or VV at codon 129, and show linear plaque formation similar to that of the kuru prion, located mainly in the basal ganglia and cerebellum. Type 4 PrPSc (MM)was associated mainly with the vCJD prion, which evidence points toward hereditary transmission of the isoform mutation. Incubation periods of different prion strains also differ, with Types 1 and 3 PrpSc having the shortest disease length (around 3-10 months). Homozygosity seems to correspond to shorter duration of illness, but the age of onset of symptoms is unrelated.[6]

Prion strains 1-4 can also be identified by differential glycosylation patterns upon partial digestion by proteinase K (shown in Figure 4). Types 1 and 2 showed similar percentages of unglycosylated, monoglycosylated, and diglycosylated substrate, while types 3 MM, 3 MV, 3 VV and type 4 MM showed very distinct patterns. Additionally, the ability of the N-terminal region of PrPSc to bind copper ions may play a role in differential classifications between types 1 and 2 PrPSc.[6]

A unique form of CJD, termed vCJD, was discovered in 1996 in Britain and is believed to be an acquired prion disease, a possible result of ingestion of brain matter from BSE-infected cattle. Symptoms are slightly different, beginning with behavioral changes, depression, anxiety, and sensory paraesthesia or dysaesthesia, and only then progressing to ataxia, progressive dementia, and myoclonus, which occur earlier in sporadic CJD. The disease duration is usually 1 to 3 years, slightly longer than in sporadic CJD as well as being found in additional areas of the body, including the tonsils, spleen, and lymph nodes.[5]
Acquired Propagation: Kuru
Kuru is an acquired prion disease symptomatically divergent from all forms of CJD. In documented kuru cases, severe dementia did not become apparent until much later after onset of the disease. Instead, it is characterized by radical behavioral changes (alternating bouts of depression and inappropriate laughter), joint pain, and loss of muscle function. Kuru is prevalent in the Fore linguistic group of the Eastern Highlands in Papua New Guinea. Members of these societies classically participated in ceremonies in which practically the entire body of deceased relatives would be feasted upon as a sign of honor and respect. Records of these ceremonies indicate that while men took choice cuts of the human flesh, mainly women and children ate the internal organs such as the brain, which is consistent with a statistically higher proportion of the 2700 documented cases of kuru being found in women and grown children. Once the source of the kuru disease was discovered in 1959 after anthropologically raised questions, better roads were built to allow governmental access to the region, and ritual endocannabalism ceased. Incidences of kuru have all but disappeared, with new cases only appearing in those individuals who participated in endocannabalistic ceremonies before their ban. Remarkably, the incubation of the prion PrpSc lasted from a minimum of 4 years to well over 40 years, the onset seeming to be more age-dependent than linked to any other factor. Proteinase K degradation patterns in all types of kuru prions are unique compared to all types of CJD prions, showing that different strains and mutations present in the forms of different symptoms (Figure 5).[7]

Although prion disease cannot be transmitted ordinarily through contact with skin or even blood, contact with brain tissue or spinal fluid does effectively transmit the disease. Dura mater grafts, corneal transplants, or surgical implantation of non-sterile electrodes in the brain can also transmit prion disease. It is not known how much abnormal PrpSc it is necessary to have ingested or injected to cause kuru or other acquired prion disease.[8]
Inherited Propagation: Fatal Familial Insomnia
Fatal Familial Insomnia is an autosomal dominant disease caused by mutation of the PRNP gene, located on chromosome 20.[5] Early symptoms include insomnia, bizarre phobias, headaches, panic attacks, and hallucinations, followed by recurrence of fetal reflexes, weight loss, apathy, breathing disorders, myoclonus, and eventually coma and death.[9] Patients rarely survived past their twenty-fifth year. Studies of the PrpSc found in FFI patients show a methionine at codon 129, as well as an asparagine mutation at codon 178.[10] Homozygosity of Met/Met at codon 129 is associated with quicker onset and shorter duration of disease, while heterozygosity Met/Val can often present after years of latency.[5]
Cross-species Transmission of Prions
Although normally transmission of prions between species is impossible due to differing amino acid sequences of the PrPC and PrPSc protein, there are several documented instances of just such occurrences. One of the main pieces of evidence for the protein-only hypothesis is that when a mutated PrPSc is inoculated into a wild-type member of the same species, the PrPSc that forms in the recipient organism has the same amino acid sequence as the PrPC in the recipient organism, not the PrPSc in the donor organism. However, if homology is close enough for PrPSc to retain activity and reconfigure wild type PrPC in the recipient species, that strain of PrPSc can cross the transmission barrier.[4]
This has been documented in yeast, in a 2005 study where researchers used a synthetic form of the S. cerevisiae prion Sup35 to infect a distantly phylogenetically related and highly divergent species of yeast, C. albicans. The introduction of Sup35 into the recipient yeast resulted in an conformationally adapted unique conformation of prion, which was then removed and inoculated back into the donor yeast re-cross the species transmission barrier. These findings not only support the 'adaptation' hypothesis of prion formation, but support the theory that conformation is the primary determinant in cross-species prion transmission, while amino acid sequence affects the specificity by allowing and prohibiting differential amyloid conformations.[11]
An area of recent interest was the hypothetical transmission of BSE, or 'mad cow' disease, to humans through consumption of beef from infected cattle. It is believed that BSE originated from scrapie-infected sheep matter in cattle feed, and hypothesized that vCJD is a result of the same process in humans. In the United Kingdom, fear of this method of prion disease transmission led to the formation of the CJD Surveillance Unit, as well as the passage of legislation to regulate the quality of beef and test for presence of BSE in cattle feed and meat intended for human consumption. Other theories have been presented, such as ingestion of droppings of mice who had been themselves exposed by eating BSE-infected cattle feed[13]. Some believe that a third isoform of the prion protein exists, a soluble PrP, whose buildup in the cytosol as a result of proteasome inhibition is the cause of neurotoxicity.[15] Still more scientists, who doubt the existence of prions as infectious agents at all, are convinced that mad cow is a result of the bacteria Mycobacterium bovis, which causes tuberculosis in cows and humans, and that methods used to prepare prion solutions and systems simply destroy the traces of the bacteria.[14]
In addition to infectious prion transmission from one species to another, the theory that prions may be carried between members of the same species by a vector has been an important subject of research. PrPSc present in the skin in keratinocytes and fibroblasts may be ingested by ectoparasties, and homogeneates of these prion-infected ectoparasites, such as Diptera (flies whose larvae burrow beneath mammalian skin in an invasion known as myiasis) and the mites Lepidoglyphus destructor and Acarus farris have been shown to be able to spread scrapie in sheep and mice.[2]
Conclusion
What importance does prion propagation have to the field of microbiology?
Microbes are defined as living organisms that require a microscope to be seen; however, the jury is still out on whether even viruses count as living organisms, despite their classification as microbes. Other microbes include microscopic eukaryotes, bacteria, and archaea. Can prions, viroid-like particles with no other molecular parts, protective coats, or nucleic acids be classified as microbes? They represent an even smaller, simpler form of pathogenic material, that can propagate itself, perhaps without the aid of any other proteins, enzymes, or cofactors. If prions are indeed the sole infectious agent responsible for their own propagation, many of the definitions and assumptions we hold about pathogens and disease would be rendered null and void.
The prion hypothesis presents even greater problems to the whole field of biology. The fundamental beliefs that DNA is transcribed to RNA which is translated to protein could be detrimentally altered, if future evidence can undeniably prove that PrPSc is propagated from other PrPSc. Future research is most definitely merited on the subject of prions. While some continue to search for elusive links to nucleic acids and virus particles, others struggle to determine the specific mechanism by which one PrPSc protein might bind and change the conformation of another, thus documenting an entirely unprecedented occurrence - the retaining of information in a vehicle outside DNA and RNA, and solely in a protein conformation.
References
Edited by Ken Farabaugh, a student of Joan Slonczewski for BIOL 238 Microbiology, 2009, Kenyon College.
