Pseudomonas putida’s Role In The Bioremediation Of Plastic
Overview
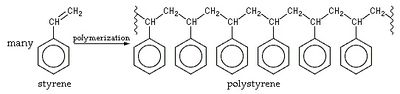
With the rapid industrial development in recent decades, hazardous substance emission is causing more concern. One of the most common materials used in synthetic plastic and rubber is polystyrene, commonly known as styrofoam. This rigid polymer is made of an aromatic monomer phenylethene or styrene. Polystyrene is found to be very recalcitrant and persistent in the environment and the inhalation of polystyrene in the air could cause problems in the central nervous system [1]. The production of polystyrene is also found to be able to cause air pollution by altering the composition of the stratosphere and the troposphere depending on the process used [2]. Therefore it is of great interest to study microorganisms capable of the degradation of styrene such as Pseudomonas putida.
Pseudomonas putida
Domain: Bacteria
Phylum: Proteobacteria
Class: Gamma proteobacteria
Order: Pseudomonadales
Family: Pseudomonadaceae
Genus: Pseudomonas
Species: Pseudomonas putida
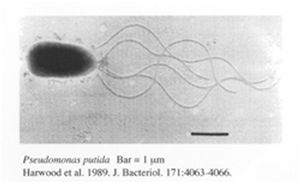
Pseudomonas putida is a gram-negative, rod-shaped bacteria which undergoes aerobic metabolism. It is normally found in terrestrial and aquatic environments where oxygen is abundant. P.putida is well known for being one of the most metabolically diverse organisms capable of breaking down both natural organic molecules such as vanillin, limonene and camphor [5] as well as industrial compounds such as toluene and styrene [4]. There is great interest in the genomic analysis of P.putida by a joint genome sequencing project by several institutes from the U.S and Germany with one of the most well studied strains being the P.putida K2440 strain. The genome of this particular strain contains 6181 kbp with 5420 ORFs. Out of all these ORFs, 3571 ORFs were assigned putative roles through comparisons against a non-redundant protein database [6]. Although these ORFs may not all be translated, the abundance of ORFs suggest that the bacteria is capable of various metabolic activities. In addition, to achieve the metabolic versatility of the bacteria requires the ability to cope with numerous abiotic stresses [7]. This is further supported by the genomic analysis of KT2440 where an unusually ample amount of transporters, oxidoreductases, mono-oxygenases, dioxygenases, ferredoxins, sulphur metabolism proteins, dehydrogenases, and cytochromes are found to be encoded in the genome [6]. Aside from P. putida, several other bacteria are also being looked into for bioremediation purposes.
Properties of Polystyrene
Huge amounts of potentially toxic styrene are produced each year, making the removal of the pollutant from the environment of critical importance. There are few microbes capable of metabolizing styrene as a sole source of carbon; these bacteria are able to transform the recalcitrant and persistent styrene into another molecule that’s easier to break down such as polyhydroxyalkanoate or PHA [10]. Styrene is the simplest alkenyl benzene. It is colourless, oil-like and not very soluble in water. The polymerization of styrene into polystyrene is a spontaneous process that can occur at room temperature [9]. Though less toxic than benzene or polycyclic aromatic hydrocarbons, styrene is still potentially carcinogenic. It also affects the central nervous system if inhaled. Chronic exposure to styrene could result in memory loss, difficulties in concentration and learning, and cancer [11]. Most industries deal with styrene pollutants through combustion or land injection. Both methods are flawed as soil styrene rapidly degrades at first but starts to persist subsequently [13]. It is also observed that aerosol styrene created from combustion is capable of damaging the ozone by reacting with O3 to form highly unstable and reactive ozonide [3][9].
Bioremediation of Styrene
It is suggested that P.putida has relatively relaxed substrate specificity; this is further supported by a study showing that when a strain of P.putida is isolated from conditions where isopropylbenzene is normally the only carbon source, it is able to grow using alternate aromatic source [14].
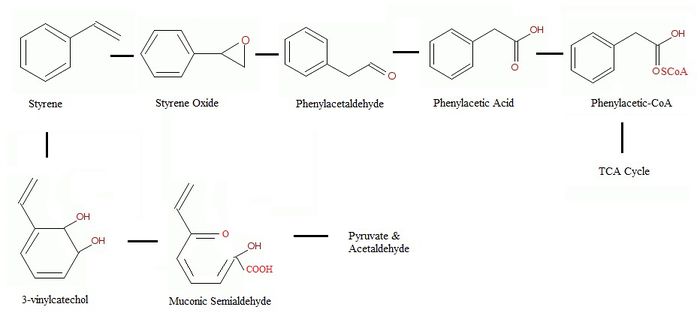
P.putida is also found to be capable of biodegrading styrene in two different pathways via two modes of initial attack of styrene from the isolation of [15]. The isolation of two different oxidation products, 2-phenyl-2-propen-1-nol and 1,2-dihydroxy-3-isopropenyl-3-cyclohexene suggest different initial steps of styrene metabolism by P.putida [14]. The first being oxidation of the vinyl side chain as the predominant pathway under aerobic conditions; the second involves the direct oxidation of the aromatic ring [9][12][14][15][16].
Side Chain Pathway
The side chain oxidation forms several intermediates that all convert to phenylacetic acid (PAA), this step is also called the “upper pathway” of the styrene metabolism [9][12]. The step after the production of phenylacetic acid is named the “lower pathway” of the styrene metabolism, which involves the production of phenylacetyl-CoA [7].
The first step of the upper pathway involves the conversion of styrene to styrene oxide, this is done by a monooxygenase. Styrene oxide is then isomerised into phenylacetaldehyde. Finally, phenylacetaldehyde would be oxidized to become phenylacetic acid [9][12]. In the lower pathway, phenylacetic acid will be joined to CoA to make phenylacetyl-CoA by a ligase. This molecule is then able to be cleaved into acetyl CoA which can enter the TCA cycle [9]. The degradation of the pathway is not linear and more branches exist based on the availability of nutrient [10].
Aromatic Ring Pathway
The direct attack on the aromatic ring results in styrene cis-glycol followed by 3-vinylcatechol formation [9][17]. Then 3-vinylcatechol will be cleaved by several enzymes such as catechol 2,3-dioxygenase, semialdehyde hydrolase, hydratase and aldolase to become a muconic semialdehyde [17]. The muconic semialdehyde will then be metabolized to pyruvate and acetaldehyde to be used in further metabolic pathways [9].
References
(1) Welp, E., Kogevinas, M., Andersen, A., Bellander, T., et al. “Exposure to Styrene and Mortality from Nervous System Diseases and Mental Disorders.” Am. J. Epidemiol. 1996, 144(7), 623-633.
(2) Przybulewska, K., Wieczorek, A. and Nowak, A. “Isolation of Microorganisms Capable of Styrene Degradation.” Polish J. of Environ. 2006, 15(5), 777-783.
(3) Hisatsune, I.C., Shinoda, K., Heicklen, J. 1978. “Low-temperature infrared studies of the styrene-ozone reaction, formation of an unusual ozonide.” J. Am. Chem. Soc. 1978, 101(10), 2524-2526.
(4) Marques, S., Ramos, J.L., “Transcriptional control of the Pseudomonas putida TOL plasmid catabolic pathways.” J. Mol. Microbiol. 1993, 9(5), 923-929.
(5) Wackett, L.P. “Pseudomonas putida – a versatile biocatalyst.” Nat. Biotech. 2003, 21(2), 136-138.
(6) Nelson, K., Weinel, C., Fraser, C., et al. “Complete genome sequence and comparative analysis of the metabolically versatile Pseudomonas putida KT2440.” Appl. Environ. Microbiol. 2002, 4(12), 799-808.
(7) Reva, O.N., Weinel, C., Weinel, M., et al. “Functional genomics of stress response in Pseudomonas putida KT2440.” J. Bacteriol. 2006, 188(11), 4079-4092.
(8) Puchalka, J., Oberhardt, M.A., Godinho, M., et al. “Genome-scale reconstruction and analysis of the Pseudomonas putida KT2440 metabolic network facilitates applications in biotechnology.” PLoS. Comput. Biol. 2008, 4(10), 1-18.
(9) Mooney, A., Ward, P.G., O’Connor, K.E. “Microbial degradation of styrene: biochemistry, molecular genetics, and perspectives for biotechnological applications.” Appl. Microbiol. Biotechnol. 2006, 71(1), 1-10.
(10) Ward, P.G., de Roo, G., O’Connor, K.E. “Accumulation of polyhydroxyalkanoate from styrene and phenylacetic acid by Pseudomonas putida CA-3.” Appl. Environ. Microbiol. 2005, 71(4), 2046-2052.
(11) Gibbs, B.F., Mulligan, C.N. “Styrene toxicity: an ecotoxicological assessment.” Exotox. Environ. Safe. 1997, 38(3), 181-194.
(12) O’Connor, K., Duetz, W., Wind, B., et al. “The effect of nutrient limitation of styrene metabolism in Pseudomonas putida CA-3.” Appl. Environ. Microbiol. 1996, 62(10), 3594-3599.
(13) Fu, M.H. and Alexander, M. “Biodegradation of styrene in waterlogged soils and aquifer solids.” Soil Science. 1996, 161(12), 846-851.
(14) Bestetti, G., Galli, E., Benigni, C., et al. “Biotransformation of styrenes by a Pseudomonas putida.” Appl. Microbiol. Biotechnol. 1989, 30(3), 252-256.
(15) Omori, T., Jigami, Y., Minoda, Y. “Microbial oxidation of a-methylstyrene and B-methylstyrene.” Agr. Biol. Chem. 1974, 38, 409-415.
(16) Jigami, Y., Kawasaki, Y., Omori, T., et al. “Coexistence of different pathways in the metabolism of N-propyl-benzene by Pseudomonas sp.” Appl. Environ. Microbiol. 1979, 38(5), 783-788.
(17) Warhurst, A. M., Clarke, K. F., Hill, R. A., et al. “Metabolism of styrene by Rhodococcus rhodochrous.” Appl. Environ. Microbiol. 1994, 60(4), 1137-1145.