Pseudomonas putida alesha & gwen: Difference between revisions
| (32 intermediate revisions by the same user not shown) | |||
| Line 20: | Line 20: | ||
''Pseudomonas putida'' | ''Pseudomonas putida'' | ||
{| | {| | ||
| Line 30: | Line 31: | ||
[[File:Collection_site_spring.jpeg|200px|thumb|left|alt=Collection site of soil sample from which Pseudomonas putida was isolated.|Collection site of P. putida in spring.]] | [[File:Collection_site_spring.jpeg|200px|thumb|left|alt=Collection site of soil sample from which Pseudomonas putida was isolated.|Collection site of P. putida in spring.]] | ||
The organism ''Pseudomonas putida'' was collected and isolated from a 3 Tbsp soil sample gathered at the Dove Springs District Park at 5801 Ainez Drive in Austin, Texas, at a location with the coordinates of Latitude: 30.184271 and Longitude: -97.736561. The collection point lay within a wooded area between the baseball field and Williamson Creek, located approximately 50 to 60 feet West of the creek and approximately 30 feet Southeast of the nearest walking trail. | |||
The organism Pseudomonas putida was collected and isolated from a 3 Tbsp soil sample gathered at the Dove Springs District Park at 5801 Ainez Drive in Austin, Texas, at a location with the coordinates of Latitude: 30.184271 and Longitude: -97.736561. The collection point lay within a wooded area between the baseball field and Williamson Creek, located approximately 50 to 60 feet West of the creek and approximately 30 feet Southeast of the nearest walking trail. | |||
The soil sample collection occurred on January 28, 2016. The temperature was 61°F with 39% humidity, no rainfall in the prior 24 hours, and solar radiation of 17.35 MJm2. Soil was collected from the surface to a depth of 1 inch. The collection location is fully shaded in summer and partly shaded in winter with low foot traffic. | The soil sample collection occurred on January 28, 2016. The temperature was 61°F with 39% humidity, no rainfall in the prior 24 hours, and solar radiation of 17.35 MJm2. Soil was collected from the surface to a depth of 1 inch. The collection location is fully shaded in summer and partly shaded in winter with low foot traffic. | ||
| Line 38: | Line 37: | ||
According to the USDA Natural Resources Conservation Service Web Soil Survey, the soil in the location of collection consists of Oakalla silty clay loam, with 0-2% slopes, and is frequently flooded. | According to the USDA Natural Resources Conservation Service Web Soil Survey, the soil in the location of collection consists of Oakalla silty clay loam, with 0-2% slopes, and is frequently flooded. | ||
Pseudomonas putida is a common organism found all over the world in soil and freshwater environments.[3] | ''Pseudomonas putida'' is a common organism found all over the world in soil and freshwater environments.[3] | ||
==Description and Significance== | ==Description and Significance== | ||
| Line 44: | Line 43: | ||
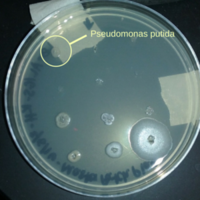
[[File:Master_patch_plate_with_Pseudomonas_putida.png|200px|thumb|right|alt=Master Patch Plate from soil sample.|Master patch plate created from collected soil samples.]] | [[File:Master_patch_plate_with_Pseudomonas_putida.png|200px|thumb|right|alt=Master Patch Plate from soil sample.|Master patch plate created from collected soil samples.]] | ||
Upon isolation and growth on an LB agar plate ''P. putida'' colonies appear mostly translucent and colorless. They are umbonate in shape and round with smooth, entire margins. Colonies appear shiny but not mucoid. | |||
Under microscopic observation after performing a Gram stain, the cellular arrangement of ''P. putida'' appears to be single rods. | |||
While none of the organisms isolated from the collected soil samples showed evidence of any kind of clearing around the colony when grown on a lawn of ''S. aureus'' or ''E. coli'', including ''P. putida'', this particular organism was selected due to the difference in colony morphology it exhibited when grown on a lawn of ''S. aureus''. Isolated colonies of ''P. putida'' appear as described above. However, when grown in the presence of ''S. aureus'', the colony of ''P. putida'' appeared dark brown in color, with a flat shape and irregular border. Although this did not suggest antimicrobial activity, it was the only isolated organism that exhibited a change in growth in the presence of another bacteria. | |||
==Genome Structure== | |||
[1] According to the Pseudomonas Genome Database the size of ''Pseudomonas putida KT2440'' genome is 6,181,863 nucleotide pairs. Of that, there is a 61.5% guanine and cytosine pairing and the other 38.5% is adenine and thymine. ''P. putida'' has a single circular chromosome. ''P. putida'' was found to share many homologous ORF's (4610/5420) with bacterium ''Pseudomonas aeruginosa''. It is the first gram negative bacteria certified as a biosafety host for cloning of foreign genes [2]. | |||
Our PCR sequencing results are as follows: | |||
Forward: Approximately 45 nucleotides were clipped from the beginning and 355 from the end to eliminate all "N" results. | |||
TGATCCAGCCATGCCGCGTGTGTGAAGAAGGTC | |||
TTCGGATTGTAAAGCACTTTAAGTTGGGAGGAAGGGCAGTAAGCTAATACCTTGCTGTTTTGACGTTACCGACAGAATAAGCACCGGCTAACTCTGTGCCAGCA | |||
GCCGCGGTAATACAGAGGGTGCAAGCGTTAATCGGAATTACTGGGCGTAAAGCGCGCGTAGGTGGTTCGTTAAGTTGGATGTGAAAGCCCCGGGCTCAACCTGGGAACTGCATCCAAAACTGGCGAGCTAGAGTACGGTAGA | |||
GGGTGGTGGAATTTCCTGTGTAGCGGTGAAATGCGTAGATATATGAAGGAACACCACTGTTCAAGGCGACCACCTGGACTGATACTGACACTGAGGTG | |||
Reverse: Approximately 13 nucleotides were clipped from the beginning and 23 from the end to eliminate all "N" results. | |||
GCACCTGTGTCAGAGTTCCCGAAGGCACCAATCCATCTCTGGAAAGTTCTCTGCATGTCAAGGCCTGGTAAGGTTCTTCGCGTTGCTTCGAATTAAACCACATGCTCCACCGCTTGTGCGGGCCCCCGTCAATTCAT | |||
TTGAGTTTTAACCTTGCGGCCGTACTCCCCAGGCGGTCAACTTAATGCGTTAGCTGCGCCACTAAAATCTCAAGGATTCCAACGGCTAGTTGACATCGTTTACGGCGTGGACTACCAGGGTATCTAATCCTGTTTGCTCCCC | |||
ACGCTTTCGCACCTCAGTGTCAGTATCAGTCCAGGTGGTCGCCTTCGCCACTGGTGTTCCTTCCTATATCTACGCATTTCACCGCTACACAGGAAATTCCACCACCCTCTACCGTACTCTAGCTCGCCAGTTTTGGATGCAG | |||
TTCCCAGGTTGAGCCCGGGGCTTTCACATCCAACTTAACGAACCACCTACGCGCGCTTTACGCCCAGTAATTCCGATTAACGCTTGCACCCTCTGTATTACCGCGGCTGCTGGCACAGAGTTAGCCGGTGCTTATTCTGTCG | |||
GTAACGTCAAAACAGCAAGGTATTAACTTACTGCCCTTCCTCCCAACTTAAAGTGCTTTACAATCCGAAGACCTTCTTCACACACGCGGCATGGCTGGATCANGCTTTCGCCCATTGTCCAATATTCCCCACTGCTGCCACC | |||
CGTAGG | |||
This was then put into a BLAST searching database and we found our results to match the closest with ''Pseudomonas putida''. | |||
==Cell Structure, Metabolism and Life Cylce== | ==Cell Structure, Metabolism and Life Cylce== | ||
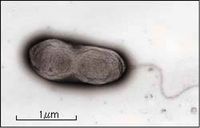
[[File:pputida.jpg|200px|thumb|left|alt=|A single prokaryotic, flagellated "P. putida F1" organism. [10]]] | |||
''Pseudomonas putida'' is a aerobic, gram negative, rod shaped bacteria. It is nonsporeforming and is motile by one or more flagella. This organism grew well on the TSA agar, LB broth and LB agar but was otherwise slow to grow in other mediums used in lab. | ''Pseudomonas putida'' is a aerobic, gram negative, rod shaped bacteria. It is nonsporeforming and is motile by one or more flagella. This organism grew well on the TSA agar, LB broth and LB agar but was otherwise slow to grow in other mediums used in lab. | ||
Although no tests were performed to prove a terminal electron acceptor, | Although no tests were performed to prove a terminal electron acceptor, ''P. putida'' does reduce nitrate to nitrite; gaining an electron in the process. | ||
==Physiology and Pathogenesis== | ==Physiology and Pathogenesis== | ||
Although our lab tests indicated that P. putida has few defining biochemical characteristics, positive results from the citrate test indicates this organism can produce citrate permease to ingest and use citrate as a sole carbon source. Additionally, further testing revealed that P. putida can produce arginine decarboxylase to faciliate decarboxylation of arginine. Finally, an alpha-positive result on the blood agar test indicated that P. putida can cause partial hemolysis of red blood cells. All other test results were negative. | Although our lab tests indicated that ''P. putida'' has few defining biochemical characteristics, positive results from the citrate test indicates this organism can produce citrate permease to ingest and use citrate as a sole carbon source. Additionally, further testing revealed that ''P. putida'' can produce arginine decarboxylase to faciliate decarboxylation of arginine. Finally, an alpha-positive result on the blood agar test indicated that ''P. putida'' can cause partial hemolysis of red blood cells. All other test results were negative. | ||
''P. putida'' is well known as a bioremediator.[9] In recent research this organism has shown to be quite effective in the breakdown of hydrocarbons and is in trials to be used in large amounts to clean up many organic toxins. The only substances ''P. putida'' has shown to not be able to break down are Teflon, Styrofoam and organic products containing only a single hydrogen atom.[3] | |||
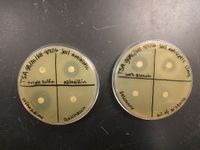
P. putida | [[File:AB_and_antiseptic_susceptibility.jpeg|200px|thumb|right|alt=Antibiotic and antiseptic susceptibility of P. putida.|Antibiotic and antiseptic susceptibility of P. putida.]] | ||
This organism was initially feared to be pathogenic because of the close comparison to ''Pseudomonas aeruginosa'', a disease-causing bacteria also in the ''Pseudomonas'' genus. This was proved to be false when scientist learned that although ''P. putida'' is a mesophile, its preferred temperature for growth is 35ºC, with humans averaging at 37ºC.[3] However, nosocomial infections of ''P. putida'' have been documented since the 1980's, particularly among the immunocompromised.[4] Several strains have demonstrated pathogenicity but the HB3267 showed particular virulence, infecting human tissues, other mammalian tissues, and even insect larvae.[5] More recently, one of the infective strains has been found to demonstrate resistance to multiple antibiotics. The resistant genes were found on chromosomes and on the pPC9 plasmid, and were believed to have come from human pathogens as well as environmental microorganisms.[6] However, not all strains show this level of resistance and tests we completed in lab indicated that our isolated organism was susceptible to azlocillin and ceftazidime. | |||
Symptoms of infections caused by ''P. putida'' ranged from phlebitis and cellulitis to gangrene.[7] Although the organism is capable of creating a biofilm, a 2012 study showed that in the KT2440 strain the biofilm showed susceptibility to cassia oil, an essential oil extracted from ''Cinnamomum aromaticum''.[8] Additionally, the susceptibility tests we completed indicated that the organism was susceptible to clove oil. | |||
==References== | |||
[http://www.pseudomonas.com/strain/show/110/ [1]] Winsor GL, Griffiths EJ, Lo R, Dhillon BK, Shay JA, Brinkman FS (2016). Pseudomonas Genome DB. The Brinkman Lab at Simon Fraser University. n.d. Web. 20 April 2016. | |||
[ | [http://www.ncbi.nlm.nih.gov/pubmed/12534463/ [2]] Nelson KE, Weinel C, Paulsen IT, Dodson RJ, Hilbert H, et al. (2002) Complete genome sequence and comparative analysis of the metabolically versatile Pseudomonas putida KT2440. Environ Microbiol 4: 799–808. | ||
[ | [http://web.mst.edu/~microbio/bio221_2007/P_putida.htm [3]] Hamilton, Kris n.d. Pseudomonas putida. Missouri University of Science and Technology. 4 May 2016. | ||
[ | [http://www.ncbi.nlm.nih.gov/pubmed/3605136/ [4]] Anaissie, E, et al. (1987) Pseudomonas putida. Newly recognized pathogen in patients with cancer. American Journal of Medicine 82(6):1191-4. | ||
[ | [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC4548156/ [5]] Fernandez, Matilde et al. (2015) Analysis of the pathogenic potential of nosocomial Pseudomonas putida strains. Frontiers in Microbiology 6:871. | ||
[ | [http://www.ncbi.nlm.nih.gov/pubmed/24465371/ [6]] Molina, Lazaro et al. (2014) Antibiotic Resistance Determinants in a Pseudomonas putida Strain Isolated from a Hospital. PLoS One 9(1): e81604. | ||
[ | [http://www.ncbi.nlm.nih.gov/pubmed/25722620/ [7]] Lugito, Hardjo, NP et al. (2015) Diabetic Foot Gangrene Patient with Multi-drug Resistant Pseudomonas Putida Infection in Karawaci District, Indonesia. Journal of Global Infectious Diseases 7(1):37-9. | ||
[ | [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3346404/ [8]] Kavanaugh, Nicole L. and Ribbeck, Katharina. (2012) Selected Antimicrobial Essential Oils Eradicate Pseudomonas spp. and Staphylococcus aureus Biofilms. Applied and Environmental Microbiology 78(11): 4057–4061. | ||
[ | [http://www.sciencedirect.com/science/article/pii/S0964830599000487/ [9]] Raghavan, P.U.M. and Vivekanandan, M. (1999) Bioremediation of oil-spilled sites through seeding of naturally adapted Pseudomonas putida. International Biodeterioration and Biodegration 44(1):29-32. | ||
[ | [http://genome.jgi.doe.gov/psepu/psepu.home.html [10]] n.p. (2016) The Regents of the University of California. Genome Portal- Pseudomonas putida F1. | ||
==Author== | ==Author== | ||
Page authored by Alesha Larkins and Gwen Schurmann, | Page authored by Alesha Larkins and Gwen Schurmann, students of Prof. Kristine Hollingsworth at Austin Community College. | ||
<!-- Do not remove this line-->[[Category:Pages edited by students of Kristine Hollingsworth at Austin Community College]] | <!-- Do not remove this line-->[[Category:Pages edited by students of Kristine Hollingsworth at Austin Community College]] | ||
Latest revision as of 19:26, 6 May 2016
Classification
Domain; Phylum; Class; Order; family [Others may be used. Use NCBI link to find]
Domain: Bacteria
Phylum: Proteobacteria
Class: Gammaproteobacteria
Order: Pseudomonadales
Family: Pseudomonadaceae
Genus: Pseudomonas
Species
Pseudomonas putida
|
NCBI: Taxonomy |
Habitat Information
The organism Pseudomonas putida was collected and isolated from a 3 Tbsp soil sample gathered at the Dove Springs District Park at 5801 Ainez Drive in Austin, Texas, at a location with the coordinates of Latitude: 30.184271 and Longitude: -97.736561. The collection point lay within a wooded area between the baseball field and Williamson Creek, located approximately 50 to 60 feet West of the creek and approximately 30 feet Southeast of the nearest walking trail.
The soil sample collection occurred on January 28, 2016. The temperature was 61°F with 39% humidity, no rainfall in the prior 24 hours, and solar radiation of 17.35 MJm2. Soil was collected from the surface to a depth of 1 inch. The collection location is fully shaded in summer and partly shaded in winter with low foot traffic.
According to the USDA Natural Resources Conservation Service Web Soil Survey, the soil in the location of collection consists of Oakalla silty clay loam, with 0-2% slopes, and is frequently flooded.
Pseudomonas putida is a common organism found all over the world in soil and freshwater environments.[3]
Description and Significance
Upon isolation and growth on an LB agar plate P. putida colonies appear mostly translucent and colorless. They are umbonate in shape and round with smooth, entire margins. Colonies appear shiny but not mucoid.
Under microscopic observation after performing a Gram stain, the cellular arrangement of P. putida appears to be single rods.
While none of the organisms isolated from the collected soil samples showed evidence of any kind of clearing around the colony when grown on a lawn of S. aureus or E. coli, including P. putida, this particular organism was selected due to the difference in colony morphology it exhibited when grown on a lawn of S. aureus. Isolated colonies of P. putida appear as described above. However, when grown in the presence of S. aureus, the colony of P. putida appeared dark brown in color, with a flat shape and irregular border. Although this did not suggest antimicrobial activity, it was the only isolated organism that exhibited a change in growth in the presence of another bacteria.
Genome Structure
[1] According to the Pseudomonas Genome Database the size of Pseudomonas putida KT2440 genome is 6,181,863 nucleotide pairs. Of that, there is a 61.5% guanine and cytosine pairing and the other 38.5% is adenine and thymine. P. putida has a single circular chromosome. P. putida was found to share many homologous ORF's (4610/5420) with bacterium Pseudomonas aeruginosa. It is the first gram negative bacteria certified as a biosafety host for cloning of foreign genes [2].
Our PCR sequencing results are as follows:
Forward: Approximately 45 nucleotides were clipped from the beginning and 355 from the end to eliminate all "N" results.
TGATCCAGCCATGCCGCGTGTGTGAAGAAGGTC TTCGGATTGTAAAGCACTTTAAGTTGGGAGGAAGGGCAGTAAGCTAATACCTTGCTGTTTTGACGTTACCGACAGAATAAGCACCGGCTAACTCTGTGCCAGCA GCCGCGGTAATACAGAGGGTGCAAGCGTTAATCGGAATTACTGGGCGTAAAGCGCGCGTAGGTGGTTCGTTAAGTTGGATGTGAAAGCCCCGGGCTCAACCTGGGAACTGCATCCAAAACTGGCGAGCTAGAGTACGGTAGA GGGTGGTGGAATTTCCTGTGTAGCGGTGAAATGCGTAGATATATGAAGGAACACCACTGTTCAAGGCGACCACCTGGACTGATACTGACACTGAGGTG
Reverse: Approximately 13 nucleotides were clipped from the beginning and 23 from the end to eliminate all "N" results.
GCACCTGTGTCAGAGTTCCCGAAGGCACCAATCCATCTCTGGAAAGTTCTCTGCATGTCAAGGCCTGGTAAGGTTCTTCGCGTTGCTTCGAATTAAACCACATGCTCCACCGCTTGTGCGGGCCCCCGTCAATTCAT TTGAGTTTTAACCTTGCGGCCGTACTCCCCAGGCGGTCAACTTAATGCGTTAGCTGCGCCACTAAAATCTCAAGGATTCCAACGGCTAGTTGACATCGTTTACGGCGTGGACTACCAGGGTATCTAATCCTGTTTGCTCCCC ACGCTTTCGCACCTCAGTGTCAGTATCAGTCCAGGTGGTCGCCTTCGCCACTGGTGTTCCTTCCTATATCTACGCATTTCACCGCTACACAGGAAATTCCACCACCCTCTACCGTACTCTAGCTCGCCAGTTTTGGATGCAG TTCCCAGGTTGAGCCCGGGGCTTTCACATCCAACTTAACGAACCACCTACGCGCGCTTTACGCCCAGTAATTCCGATTAACGCTTGCACCCTCTGTATTACCGCGGCTGCTGGCACAGAGTTAGCCGGTGCTTATTCTGTCG GTAACGTCAAAACAGCAAGGTATTAACTTACTGCCCTTCCTCCCAACTTAAAGTGCTTTACAATCCGAAGACCTTCTTCACACACGCGGCATGGCTGGATCANGCTTTCGCCCATTGTCCAATATTCCCCACTGCTGCCACC CGTAGG
This was then put into a BLAST searching database and we found our results to match the closest with Pseudomonas putida.
Cell Structure, Metabolism and Life Cylce
Pseudomonas putida is a aerobic, gram negative, rod shaped bacteria. It is nonsporeforming and is motile by one or more flagella. This organism grew well on the TSA agar, LB broth and LB agar but was otherwise slow to grow in other mediums used in lab.
Although no tests were performed to prove a terminal electron acceptor, P. putida does reduce nitrate to nitrite; gaining an electron in the process.
Physiology and Pathogenesis
Although our lab tests indicated that P. putida has few defining biochemical characteristics, positive results from the citrate test indicates this organism can produce citrate permease to ingest and use citrate as a sole carbon source. Additionally, further testing revealed that P. putida can produce arginine decarboxylase to faciliate decarboxylation of arginine. Finally, an alpha-positive result on the blood agar test indicated that P. putida can cause partial hemolysis of red blood cells. All other test results were negative.
P. putida is well known as a bioremediator.[9] In recent research this organism has shown to be quite effective in the breakdown of hydrocarbons and is in trials to be used in large amounts to clean up many organic toxins. The only substances P. putida has shown to not be able to break down are Teflon, Styrofoam and organic products containing only a single hydrogen atom.[3]
This organism was initially feared to be pathogenic because of the close comparison to Pseudomonas aeruginosa, a disease-causing bacteria also in the Pseudomonas genus. This was proved to be false when scientist learned that although P. putida is a mesophile, its preferred temperature for growth is 35ºC, with humans averaging at 37ºC.[3] However, nosocomial infections of P. putida have been documented since the 1980's, particularly among the immunocompromised.[4] Several strains have demonstrated pathogenicity but the HB3267 showed particular virulence, infecting human tissues, other mammalian tissues, and even insect larvae.[5] More recently, one of the infective strains has been found to demonstrate resistance to multiple antibiotics. The resistant genes were found on chromosomes and on the pPC9 plasmid, and were believed to have come from human pathogens as well as environmental microorganisms.[6] However, not all strains show this level of resistance and tests we completed in lab indicated that our isolated organism was susceptible to azlocillin and ceftazidime.
Symptoms of infections caused by P. putida ranged from phlebitis and cellulitis to gangrene.[7] Although the organism is capable of creating a biofilm, a 2012 study showed that in the KT2440 strain the biofilm showed susceptibility to cassia oil, an essential oil extracted from Cinnamomum aromaticum.[8] Additionally, the susceptibility tests we completed indicated that the organism was susceptible to clove oil.
References
[1] Winsor GL, Griffiths EJ, Lo R, Dhillon BK, Shay JA, Brinkman FS (2016). Pseudomonas Genome DB. The Brinkman Lab at Simon Fraser University. n.d. Web. 20 April 2016.
[2] Nelson KE, Weinel C, Paulsen IT, Dodson RJ, Hilbert H, et al. (2002) Complete genome sequence and comparative analysis of the metabolically versatile Pseudomonas putida KT2440. Environ Microbiol 4: 799–808.
[3] Hamilton, Kris n.d. Pseudomonas putida. Missouri University of Science and Technology. 4 May 2016.
[4] Anaissie, E, et al. (1987) Pseudomonas putida. Newly recognized pathogen in patients with cancer. American Journal of Medicine 82(6):1191-4.
[5] Fernandez, Matilde et al. (2015) Analysis of the pathogenic potential of nosocomial Pseudomonas putida strains. Frontiers in Microbiology 6:871.
[6] Molina, Lazaro et al. (2014) Antibiotic Resistance Determinants in a Pseudomonas putida Strain Isolated from a Hospital. PLoS One 9(1): e81604.
[7] Lugito, Hardjo, NP et al. (2015) Diabetic Foot Gangrene Patient with Multi-drug Resistant Pseudomonas Putida Infection in Karawaci District, Indonesia. Journal of Global Infectious Diseases 7(1):37-9.
[8] Kavanaugh, Nicole L. and Ribbeck, Katharina. (2012) Selected Antimicrobial Essential Oils Eradicate Pseudomonas spp. and Staphylococcus aureus Biofilms. Applied and Environmental Microbiology 78(11): 4057–4061.
[9] Raghavan, P.U.M. and Vivekanandan, M. (1999) Bioremediation of oil-spilled sites through seeding of naturally adapted Pseudomonas putida. International Biodeterioration and Biodegration 44(1):29-32.
[10] n.p. (2016) The Regents of the University of California. Genome Portal- Pseudomonas putida F1.
Author
Page authored by Alesha Larkins and Gwen Schurmann, students of Prof. Kristine Hollingsworth at Austin Community College.