Chest Port Microbial Infections: Difference between revisions
| (2 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
<!-- Do not edit this line-->{{Curated}} | <!-- Do not edit this line-->{{Curated}} | ||
==Introduction== | ==Introduction== | ||
[[Image:Chestport.jpg|thumb| | [[Image:Chestport.jpg|thumb|300px|right|<b>Figure 1.</b> Example of Chest Port Placement. Diagram from [https://www.fairview.org/healthlibrary/Article/40934 Fairview Hospital].]] | ||
By Hannah Lorico Hertz | By Hannah Lorico Hertz | ||
| Line 29: | Line 29: | ||
As foreign materials, such as catheters, are placed into the body, they become covered by host ECM components and so, as chest ports are inserted in the body, the ports are coated by EMC components such as fibrinogen, vitronectin, collagen, and fibronectin. Research has shown that proteins with ECM-binding activity expressed by <i>S. epidermidis</i> can play a significant role in microbial biofilm initiation of a port infection. Proteins such as AltE bind to vitronectin, lipase GehD to collagen<ref name=Bowden></ref> | As foreign materials, such as catheters, are placed into the body, they become covered by host ECM components and so, as chest ports are inserted in the body, the ports are coated by EMC components such as fibrinogen, vitronectin, collagen, and fibronectin. Research has shown that proteins with ECM-binding activity expressed by <i>S. epidermidis</i> can play a significant role in microbial biofilm initiation of a port infection. Proteins such as AltE bind to vitronectin, lipase GehD to collagen<ref name=Bowden></ref>, and surface component serine-asparatate repeat (Sdr) proteins SdrF, SdrG, and SdrH<ref>[https://www.ncbi.nlm.nih.gov/pubmed/10878118/ McCrea K. W., Hartford O., Davis S., Eidhin D. N., Lina G., Speziale P., et al. "The serine-aspartate repeat (Sdr) protein family in <i>Staphylococcus epidermidis</i>." <b>(2000)</b> <i>Microbiology</i> 146: 1535–1546.]</ref> to collagen I<ref>[https://www.ncbi.nlm.nih.gov/pubmed/16544251/ Arrecubieta C., Asai T., Bayern M., Loughman A., Fitzgerald J. R., Shelton C. E., Baron, H.M., Dang, N.C., Deng, M.C., Naka,Y., Foster, T.J., Lowy, F.D. "The role of <i>Staphylococcus aureus</i> adhesins in the pathogenesis of ventricular assist device-related infections." <b>(2006)</b> <i>J. Infect. Dis.</i> 193:1109–1119.] </ref>, fibrinogen, and fibrinogen respectively. The research outlined above is important because engineers might test different chest port surfaces to which <i>S. epidermidis</i> cells can not adhere. If able to develop a chest port where <i>S. epidermidis</i> cannot bind, biofilm growth would be severely hindered and chest port infection incidence decreased. [[Image:PIA.gif|thumb|350px|right|<b>Figure 5.</b> PIA synthesis via membrane proteins <i>Ica</i>A, <i>Ica</i>D, <i>Ica</i>C and protein <i>Ica</i>B. Diagram generated by the Department of Microbiology and Immunology, Botucatu Biosciences Institute, [http://www.scielo.br/scielo.php?script=sci_arttext&pid=S1678-91992008000400003/ São Paulo State University, Brazil.].]] | ||
, and surface component serine-asparatate repeat (Sdr) proteins SdrF, SdrG, and SdrH<ref>[https://www.ncbi.nlm.nih.gov/pubmed/10878118/ McCrea K. W., Hartford O., Davis S., Eidhin D. N., Lina G., Speziale P., et al. "The serine-aspartate repeat (Sdr) protein family in <i>Staphylococcus epidermidis</i>." <b>(2000)</b> <i>Microbiology</i> 146: 1535–1546.]</ref> to collagen I<ref>[https://www.ncbi.nlm.nih.gov/pubmed/16544251/ Arrecubieta C., Asai T., Bayern M., Loughman A., Fitzgerald J. R., Shelton C. E., Baron, H.M., Dang, N.C., Deng, M.C., Naka,Y., Foster, T.J., Lowy, F.D. "The role of <i>Staphylococcus aureus</i> adhesins in the pathogenesis of ventricular assist device-related infections." <b>(2006)</b> <i>J. Infect. Dis.</i> 193:1109–1119.] </ref>, fibrinogen, and fibrinogen respectively. The research outlined above is important because engineers might test different chest port surfaces to which <i>S. epidermidis</i> cells can not adhere. If able to develop a chest port where <i>S. epidermidis</i> cannot bind, biofilm growth would be severely hindered and chest port infection incidence decreased. [[Image:PIA.gif|thumb|350px|right|<b>Figure 5.</b> PIA synthesis via membrane proteins <i>Ica</i>A, <i>Ica</i>D, <i>Ica</i>C and protein <i>Ica</i>B. Diagram generated by the Department of Microbiology and Immunology, Botucatu Biosciences Institute, [http://www.scielo.br/scielo.php?script=sci_arttext&pid=S1678-91992008000400003/ São Paulo State University, Brazil.].]] | |||
| Line 37: | Line 36: | ||
[[Image:ICAloci.png|thumb|300px|right|<b>Table 1.</b> Biofilm formation and PIA synthesis for <i>S epidermidis</i> <i>ica</i> mutant strains grown in TSB<sub>BBL</sub> and TSB<sub>Oxoid</sub> media. Table from | [[Image:ICAloci.png|thumb|300px|right|<b>Table 1.</b> Biofilm formation and PIA synthesis for <i>S epidermidis</i> <i>ica</i> mutant strains grown in TSB<sub>BBL</sub> and TSB<sub>Oxoid</sub> media. Table from [http://iai.asm.org/content/68/7/3799/T3.expansion.html/ Mack et al. (2000).].]]The study by Mack et al. (2000) confirmed the PIA model outlined in Figure 5. In this study, Mack et. al created <i>S. epidermidis</i> mutants that did not produce biofilms. They created two major categories of mutants: mutants with an insertion in the<i> ica</i>A gene (M13 and M24) and mutants with an insertion in the <i>ica</i>C gene (M21, M22, M23, Tn917). Mack et al. studied the phenotypic biofilm response of these mutations (Table 1). With the media TSB<sub>Oxoid</sub>, mutations in the <i>ica</i>A and <i>ica</i>C gene both completely knocked out biofilm formation in comparison to biofilm formation in wild type <i>S. epidermidis</i>. This same finding was observed with the media TSB<sub>BBL</sub>. However, the mutant strain M21 presented minimal biofilm formation in TSB<sub>BBL</sub> but not in TSB<sub>Oxoid</sub>. Because the insertion in the mutant M21 occurred at the end of the <i>ica</i>C gene, researchers suggest that a semi-functional protein is expressed and able to synthesize low levels of PIA, thus forming some biofilms. For the mutants both grown in TSB<sub>BBL</sub> and TSB<sub>Oxoid</sub>, PIA formation was not observed, with the exception of the low expression in M21 (Table 1). This suggests that the <i>ica</i>A and <i>ica</i>C gene are responsible for proteins involved in PIA synthesis. Mack et al.’s research confirms the role of the <i>ica</i> gene cluster in PIA synthesis and thus biofilm formation. <ref>[http://iai.asm.org/content/68/7/3799.full#cited-by/ Mack, D., Rohde, H., Dobinsky, S., Riedewald, J., Nedelmann, M., Knobloch, J., Elsner, H. and Hubert Feucht. "Identification of Three Essential Regulatory Gene Loci Governing Expression of <i>Staphylococcus epidermidis</i> Polysaccharide Intercellular Adhesin and Biofilm Formation" <b>(2000)</b> <i>Infect. Immun.</i>. 68: 3799-3807.]</ref> | ||
Latest revision as of 19:34, 12 May 2017
Introduction

By Hannah Lorico Hertz
A chest port is an indwelling catheter connected to a reservoir, inserted under the skin of the chest, and used to administer medicines directly into a vein over a long period of time (Figure 1). Physicians frequently utilize chest ports to administer multiple cycles of chemotherapy in children because of the ease of vascular access and care for port maintenance. Because chest ports are inserted under the skin, the skin covering the port acts as a barrier to potentially harmful microorganisms. In comparison to an IV line, chest ports can stay in place for months at a time, can be used to collect blood samples without needles, and have a lower risk of infection over time. Although chest port infections are not as common as other external catheter infections, the most significant complication during chemotherapeutic treatment via chest port catheters are microbial infections. Infections of implanted devices most commonly result from Staphylococcus epidermidis, Staphylococcus aureus, Enterococcus faecalis, Streptococcus vidrians, Klebsiella pneumonia, and Pseudomona aeruginosa.[1] Of the above microbes, S. epidermidis is the most relevant port associated pathogen. Jukes et al. estimate in the United States, 80% of hospital acquired catheter related bloodstream infections (CRBSI) are caused by the microbe S. epidermidis.[2] The treatment of these catheter-related bloodstream S. epidermidis infections costs the U.S. about $2 billion a year.[3] Thus, research on practices that decrease chest port infection incidence and basic scientific research on the mechanism of chest infections can significantly reduce the problem of chest port infections.
Researchers focus their study on the microbe S. epidermidis pathogenicity because of its high incidence in chest port infections. S. epidermidis are normally non-pathogenic and found on the human skin.[4] S. epidermidis are opportunistic pathogens--they only act as a human pathogen in individuals with compromised immune systems, immunosuppression, or chemotherapy related neutropenia.[4] Common port infections include S. epidermidis biofilm formation inside the catheter lumen.[1] Biofilms are groups of cells that form on various surfaces that produce extracellular polymeric substances (EPS) which protects the microorganisms from antibiotics.[5] Biofilm formation is threefold. S. epidermidis adhere to the catheter surface to be colonized, a microcolony forms, and S. epidermidis cells detach from a mature biofilm allowing S. epidermidis colonization on additional body sites. Once biofilms form on chest ports, the patients are treated with antibiotics, and, if the antibiotics are unsuccessful, by port excision. Approximately 5% of patients diagnosed with a microbial port infection are treated by port excision.[6] Better management of chest port microbial infections can help improve patient quality of life as they undergo treatment and avoid the stressors of port excision. Recent research explores how to best manage port microbial infections avoiding the current treatment of port excision. The literature focuses on both the mechanism of S. epidermidis pathogenicity/biofilm formation in order to develop better antibiotics and also alternate treatments for port infections.
S. epidermidis and Pathogenicity

In order to develop better treatments for chest port infections, researchers must first understand what differentiates pathogenic S. epidermidis from non-pathogenic strains. S. epidermidis is the most abundant pathogen comprising the human skin flora with, under normal conditions, a benign relationship with its host. [3] To begin understanding the hallmarks of S. epidermidis pathogenicity, researchers compared S. epidermidis to the common antibiotic resistant strains of S. aureus and found that S. epidermidis lack the enzyme coagulase, which is present in pathogenic S. aureus.[3] Thus, the enzyme coagulase does not determine pathogenicity.
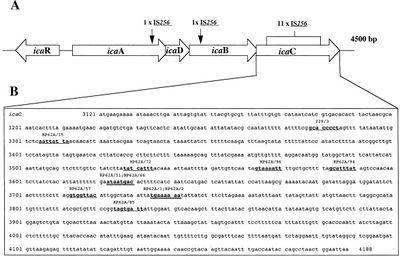
What determines pathogenic S. epidermidis strains from non-pathogenic S. epidermidis living peacefully on the human skin? Although S. epidermidis act only as pathogens in immunocompromised patients, the strains that become pathogenic in immunocompromised patients share common characteristics. One determinant of S. epidermidis pathogenicity includes a strain’s ability to form biofilms (Figure 2). S. epidermidis strains that produce biofilm are more virulent than strains which do not form biofilms.[7] Only some S. epidermidis strains, such as RP62a, form biofilms whereas other strains, such as ATCC 12228, do not form biofilms. [8] Ziebuhr et al. (1999) find the ica gene cluster (icaA, icaB, icaC, and icaD) is commonly found and highly expressed in pathogenic S. epidermidis strains isolated from catheter related infections. [9] Ica operon expression is essential for the formation of the adhesion protein PIA which promotes biofilm formation.[9] The insertion element IS 256 disrupts ica gene expression, and thus PIA formation, in non-pathogenic S. epidermidis. [9] Ziebuhr et al. study the location of IS 256 insertion in S. epidermidis which do not form biofilms in order to determine gene loci essential for S. epidermidis pathogenicity (Figure 3).[9] The authors find that for 11 out of the 13 S. epidermidis mutants that do not form biofilms, an insertion sequence (IS 256) disrupts the icaC nucleotide sequence and expression. One mutant carried the element in the icaA and the other in the icaB gene. Therefore, insertions in the icaC gene halts PIA formation and thus biofilm formation, leading to non-pathogenic S. epidermidis strains. Ziebuhr et al. also explored the excision rate of IS 256 in order to elucidate the formation of new pathogenic S. epidermidis strains. From this study, the excision rate of IS 256 was estimated to be <10-8 per cell per generation.[9] The low excision rate suggests that the introduction of new pathogenic S. epidermidis strains is relatively low.
Other regions in the S. epidermidis genome determine strain virulence. Gill et al. (2005) study regions in the biofilm forming RP62a genome responsible for S. epidermidis pathogenicity. Genes encoding for cadmium resistance and surface proteins associated with pathogenicity are found in integrated plasmids in S. epidermidis.[10] Moreover, genomic islands encode virulence factors such as multiple phenol-soluble modulins, molecules that induce the synthesis of cytokine, and the cap operon, which encodes the polyglutamate capsule.[10] Single nucleotide polymorphisms in genes encoding cell envelope proteins are also thought to affect S. epidermidis pathogenicity.
Biofilm Formation

In order to understand S. epidermidis port infections and develop better treatments, we also need to understand the process of S. epidermidis biofilm formation (Figure 4) because the most common port infections are due to S. epidermidis biofilm formation inside the catheter lumen.[1] Biofilms usually protect microbes from antibiotics. S. epidermidis biofilm formation begins with cell attachment to a surface. For port microbial infections, biofilm formation most commonly occurs on the catheter lumen. Next, S. epidermidis begin producing polysaccharides forming a protective layer surrounding the cells. The biofilm continues to grow forming mature biofilms until the cells are released into its environment, free to initiate biofilm formation on another surface (Figure 4).[11]
Biofilm formation begins as S. epidermidis adhere to the surface of the port catheter lumen. This initial attachment is essential for the establishment of port-associated infections. Attachment of S. epidermidis can occur either directly to the port polymer surface via physio-chemical interactions or indirectly by the attachment to extracellular matrix proteins (ECM) coating the surface of a port catheter. The hydrophobic cell surface of S. epidermidis due to proteins AltE and Bap/Bhp mediate the surface adhesion of the microbes to the port hydrophobic surface.[3] The autolysin AtlE is especially essential for S. epidermidis attachment to polystyrene (plastic).[12] AltE interacts to the surface of the port catheter lumen by hydrophobic interactions, however, further research on AltE mechanism of attachment is needed to eliminate the possibility that AltE plays only an indirect role in attachment.[11] Because the amino acid sequence of AltE is 61% identical to the AltL gene of Staphylococcus aureus, Heilmann et al. (1997) were able to identify two domains of interest for AltE mediation of attachment to plastic surfaces: a 60kDa amidase and a 52kDa glucosaminidase domain. [11] To test if the 60 kDa AltE domain plays a role in S. epidermidis polystyrene attachment, Heilmann et al. (1997) incubated wild type S. epidermidis with an anti-60 kDa antiserum. If the 60 kDa amidase mediates primary attachment to polystyrene, then the researchers expected to see an inhibitory effect on attachment in the initial adherence assay. As expected, with the incubation of anti-60kDa antiserum, the number of wild-type cells attached to the polystyrene plates was approximately 90% reduced.[11] This suggests that the 60 kDa domain of AltE somehow mediates S. epidermidis’ initial and direct attachment to the chest port surface during biofilm formation. AltE plays a significant role in biofilm attachment; Not only does AltE play a significant role in binding to polystyrene surfaces, but also to ECM coated surfaces (specifically vitronectin).[11]
As foreign materials, such as catheters, are placed into the body, they become covered by host ECM components and so, as chest ports are inserted in the body, the ports are coated by EMC components such as fibrinogen, vitronectin, collagen, and fibronectin. Research has shown that proteins with ECM-binding activity expressed by S. epidermidis can play a significant role in microbial biofilm initiation of a port infection. Proteins such as AltE bind to vitronectin, lipase GehD to collagen[12], and surface component serine-asparatate repeat (Sdr) proteins SdrF, SdrG, and SdrH[13] to collagen I[14], fibrinogen, and fibrinogen respectively. The research outlined above is important because engineers might test different chest port surfaces to which S. epidermidis cells can not adhere. If able to develop a chest port where S. epidermidis cannot bind, biofilm growth would be severely hindered and chest port infection incidence decreased.

By studying biofilm accumulation, researchers may be able to develop biofilm inhibitors. Biofilm accumulation of S. epidermidis begins with the expression of intercellular adhesive properties. The protein, polysaccharide intercellular adhesin (PIA), is the main adhesive expressed by S. epidermidis[2] leading to cell aggregation and biofilm formation.[15] PIA is produced by the membrane proteins IcaA, IcaD, and IcaC, which are expressed by the ica locus (Figure 5).[11] PIA formation begins with the expression of IcaA and IcaD. The proteins expressed by these genes form the N-acetylglucosamine transferase. This transferase uses UDP-glucosamine (UDP-GlcNAc) to build unbranched glucosamine (GlcNAc) homopolymers with a length of about 15-20 residues. The protein expressed by the IcaC gene produces longer chains, the PIA molecule, and is also involved in PIA export. PIA can undergo editing by IcaB, which de-acetylates some of the GlcNac residues, conferring a positive charge to a formerly neutral GlcNAC when in an environment with a pH <6.[11] PIA can then function as an adhesive for S. epidermidis biofilm production. In addition to PIA, proteins Aap and Embp can also lead to biofilm assembly. Other biofilm associated proteins involved with biofilm accumulation include SesC.[16]
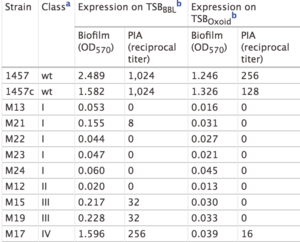
The study by Mack et al. (2000) confirmed the PIA model outlined in Figure 5. In this study, Mack et. al created S. epidermidis mutants that did not produce biofilms. They created two major categories of mutants: mutants with an insertion in the icaA gene (M13 and M24) and mutants with an insertion in the icaC gene (M21, M22, M23, Tn917). Mack et al. studied the phenotypic biofilm response of these mutations (Table 1). With the media TSBOxoid, mutations in the icaA and icaC gene both completely knocked out biofilm formation in comparison to biofilm formation in wild type S. epidermidis. This same finding was observed with the media TSBBBL. However, the mutant strain M21 presented minimal biofilm formation in TSBBBL but not in TSBOxoid. Because the insertion in the mutant M21 occurred at the end of the icaC gene, researchers suggest that a semi-functional protein is expressed and able to synthesize low levels of PIA, thus forming some biofilms. For the mutants both grown in TSBBBL and TSBOxoid, PIA formation was not observed, with the exception of the low expression in M21 (Table 1). This suggests that the icaA and icaC gene are responsible for proteins involved in PIA synthesis. Mack et al.’s research confirms the role of the ica gene cluster in PIA synthesis and thus biofilm formation. [17]
The proteins and factors involved with S. epidermidis detachment are less widely studied. However, understanding the dispersion of S. epidermidis biofilm is important in providing a holistic view of S. epidermidis biofilm formation.
The understanding of S. epidermidis pathogenicity is linked to biofilm formation. Thus, many studies have been conducted studying biofilm formation in vitro. Further research of biofilm systems in complex models is necessary to more accurately portray the in vivo infection of a chest port. The translation of the basic research on biofilm formation provides a trajectory for further chest port treatment research.
Symptoms and Diagnosis of Infection
Chest port infections most commonly occur in patients with compromised immune systems, immunosuppression, and chemotherapy related neutropenia. There are two major categories of chest port infections. The first, pocket infection, can be diagnosed by erythema, a rash caused by infection; tenderness, pain when pressure is applied to the chest port site; induration, localized hardening at the chest port site; and purulence, pus, at the chest port site.[6] The onset of a port infection can be recognized by numerous symptoms including a high fever (≥ 38.3°C or 101°F) and redness at the port site. The second, septicemia or blood poisoning due to microbes from the chest port, is more difficult to diagnose. Catheter-related septicemia is the infection of the blood originating from the port. Septicemia can often lead to sepsis, an inflammation throughout the body often forming clots that lead to organ failure.[18] In immunocompromised patients, who lack inflammatory responses, systematic catheter-related septicemia is diagnosed by exclusion because the only detectable symptom is fever and chills.[19]
The diagnostic workup of an infection includes blood cultures from the chest port catheter and a peripheral vein. These cultures are used to determine the site of infection. If the colony counts from the blood culture obtained from the chest port catheter are greater than 5-10 times of the blood culture from the peripheral blood, the physician may identify the chest port catheter as the site of infection.[20] The port may also be sent to the microbiology lab and qualitative cultures of the port reservoir contents and catheter tip compared to pinpoint the site of infection.[18]
Current Treatments

Upon the diagnosis of a port infection, doctors most commonly place the patient on broad-spectrum antibiotics. A broad spectrum antibiotic is an antibiotic that kills a vast array of gram positive and negative microorganisms. Some examples of broad spectrum antibiotics include amoxicillin, levofloxacin, gatifloxacin, streptomycin, tetracycline, and chloramphenicol.[21] The advantage of using broad spectrum antibiotics includes its coverage of multiple organisms. Furthermore, when using broad spectrum antibiotics, it is not necessary to wait to identify the specific pathogen before commencing treatment of infection. On the other hand, the use of broad spectrum antibiotics can lead to drug resistant pathogens. The arise of antibiotic resistant S. epidermidis strains makes the research on alternative chest port infection treatments vital.
The broad-spectrum antibiotic treatment of S. epidermidis port infections is problematic because S. epidermidis biofilms are resistant to many antibiotics.[3] Because S. epidermidis biofilms down-regulate many of the basic cell processes (ie. protein biosynthesis) targeted by antibiotics, antibiotics such as penicillin or quinolones have limited ability to inhibit S. epidermidis biofilm growth.[3] The limited activity of antibiotics on S. epidermidis is also due to antibiotic overuse in hospitals. [3] Hospital overuse of antibiotics has lead to increasing S. epidermidis antibiotic resistant genes. However, physicians may still utilize the antibiotic vancomycin to reduce S. epidermidis biofilm formation with success, about 80% of infections treated with antibiotics are successfully treated without port excision.[3]
If antibiotic treatment is unsuccessful, infected chest ports are removed whether or not other sites of infection are found and regardless of local signs and symptoms (Figure 6).[6] The best method of wound closure and healing (primary or secondary intention) after port removal is a highly debated process. Open drainage of the wound is the current strategy after port removal because closure can result in cellulitis or abscess if the wound is still infected (Figure 6).[6] However, open drainage is a highly involved practice. The physician must pack the wound with daily for up to two weeks, letting the wound heal by secondary intention.[6] Secondary intention is when the wound is left open to heal and close naturally. Some patients treated with open drainage require plastic surgical consultations because of unaesthetic wound healing.
After port removal and antibiotic treatment, the port is reinserted at a new site. The port may not be inserted prior to 24-72 hours in order to avoid reinfection with concurrent bacteremia.[22]
Better Treatment Methods
The current approach of antibiotic treatment then port excision is cumbersome and impractical for patients for whom venous access is critical for treatment. Thus, researchers are striving to develop alternative treatment options for microbial port infections that do not require the immediate excision of the port and replacement at a new site.
Because open drainage is time consuming, Funaki et al. (2005) studied more efficient methods to treat port microbial infections. Even if antibiotic treatment is unsuccessful, the authors argue that it is safe and preferable to close port pockets which exhibit no signs of infection. The authors argue that “all wounds contain some bacteria, all are ‘contaminated’ at some point”.[6] In fact, the study of Robson et al. (1992) determine that wounds can tolerate a contamination of up to 100,000 microbes per gram of tissues and still heal by primary intention. Primary intention is the closing of a wound with sutures. This differs from the current practice of letting the wound close by secondary intention after chest port excision due to infection. The study of Funaki et al. (2005) find that in patients with port-related septicemia who lack redness, purulence, induration, and pain at the port site, primary wound closure with antibiotic treatment also appears to be safe. This method of treatment is preferable because it simplifies the involvement of the physician follow-up wound care and provides better cosmetic results. Unfortunately, with patients who have continued redness, purulence, and induration at the port site after excision, physicians have little choice but to treat the infection with open drainage as this remains the most effective treatment method.
Caring for a Port
The best way to avoid chest port excision is to prevent infections in the first place. For example, proper sterilization technique during port placement greatly decreases the likelihood of port pocket infections. After insertion, it is also important to adopt a regular routine for port care. The caregiver must wash their hands and always sterilize the skin covering a chest port before needle insertion and treatment. Only people trained by a physician or caregiver may access a port because they are trained to use the proper technique to avoid infection. Although port care is much simpler than other catheter care due to its location under the patient’s skin, the chest port must be flushed every four weeks even when it is not in use. The chest port is also flushed after treatments and before the needle is removed. [23] Flushing a chest port is important because it prevents clot formation and blockage. A port may be flushed with either saline or heparin, a blood thinner. [23]
Conclusion
The chest port is an important device used to administer medicine over long periods of time. Chest ports are especially important for administering chemotherapy to pediatric oncology patients because patients can resume normal activity after port insertion. Furthermore, ports have a lower risk of infection. Despite the low risk of infection of ports, the primary cause of port excision is due to infection, the most prevalent infection due to S. epidermidis. An opportunistic pathogen, S. epidermidis is pathogenic only in immunocompromised patients.
The primary hallmark of S. epidermidis pathogenicity is biofilm formation. Biofilm formation begins with cell attachment. S. epidermidis cells may attach both directly to the port catheter surface or to extracellular matrix proteins coating the port surface. Biofilm accumulation begins with the production of the cell adhesion protein PIA. In fact, pathogenic strains do not contain the insertion element IS 256 which prevents PIA production. Little research has explored the factors that result in S. epidermidis biofilm detachment. Further study of detachment can lead to a better understanding of the altered pathogenic strain genomes of S. epidermidis. Furthermore, most of the studies on S. epidermidis biofilms were conducted in vitro using polystyrene to mimic the port surface. Thus, we suggest further study to explore biofilm mechanism in vivo.
There are two distinct types of port infection. The first, port pocket infections, and the second, port-related septicemia, are both treated with broad-spectrum antibiotics initially. If the antibiotics prove unsuccessful, the port is excised. Recent research suggests that wound closure, if the site does not exhibit signs of infection, is safe after excision. Wound closure is less cumbersome and more efficient than the current method of open drainage after port excision. However, the physician must make an informed decision whether or not to close an excision site based on the likelihood that there are more than 100,000 microbes per gram of tissue. It is difficult to define the parameters of a wound that does not exhibit signs of infection. Thus, the physician must balance the benefits of an efficient treatment (wound closure) versus the patient’s history and the potential ramifications if the wound is closed even if it is still infected.
References
- ↑ 1.0 1.1 1.2 Paredes, J.,Alonso-Acre, M., Schmidt, C., Valderas, D., Sedano, B., Legarda, J., Arizti, F., Gomez, E., Aguinaga, A., Del Pozo, J.L., Arana, S. "Smart central venous port for early detection of bacterial biofilm related infections" (2014) Biomed Microdevices, 16: 365.
- ↑ 2.0 2.1 Jukes, L., Mikhail, J., Bome-Mannathoko, N., Hadfield, S.J., Harris, L.G., El-Bouri, K., Davies, A.P., Mack, D. "Rapid differentiation of Staphylococcus aureus,Staphylococcus epidermidis and other coagulase-negative staphylococci and meticillin susceptibility testing directly from growth-positive blood cultures by multiplex real-time PCR" (2010) J. Med. Microbiol, 59:1456–1461
- ↑ 3.0 3.1 3.2 3.3 3.4 3.5 3.6 3.7 Otto, Michael. “Staphylococcus Epidermidis – the ‘accidental’ Pathogen.” (2009) Nature reviews. Microbiology 7.8: 555–567. PMC.
- ↑ 4.0 4.1 Buttner, H., Dietrich, M. and H. Rohde. "Structural Basis of Staphylococcus Epidermidis Biofilm Formation: Mechanisms and Molecular Interactions." (2015) Frontiers in Cellular and Infection Microbiology 5: 14. PMC.
- ↑ Donlan, Rodney M. “Biofilms: Microbial Life on Surfaces.” Emerging Infectious Diseases 8.9 (2002): 881–890. PMC. Web. 24 Apr. 2017.
- ↑ 6.0 6.1 6.2 6.3 6.4 6.5 Funaki, Brian. “Subcutaneous Chest Port Infection.” (2005). Seminars in Interventional Radiology, 22.3: 245–247. PMC.
- ↑ Deighthon, M., Borland, R., Capstick, J.A. “Virulence of Staphylococcus epidermidis in a mouse model: significance of extracellular slime.” (1996). Epidemiol Infect 117: 267–280.
- ↑ Presterl, E., Suchomel, M., Eder, M., Reichmann, S., Lassnigg, A., Graninger, W., Rotter, M. “Effects of alcohols, povidone-iodine and hydrogen peroxide on biofilms of Staphylococcus epidermidis”. (2007) Journal of Antimicrobial Chemotherapy. 60: 417-420.
- ↑ 9.0 9.1 9.2 9.3 9.4 Ziebuhr W, et al. “A novel mechanism of phase variation of virulence in Staphylococcus epidermidis: evidence for control of the polysaccharide intercellular adhesin synthesis by alternating insertion and excision of the insertion sequence element IS 256.” (1999) Mol Microbiol. 32:345–356.
- ↑ 10.0 10.1 Gill SR, Fouts DE, Archer GL, Mongodin EF, Deboy RT, Ravel J, Paulsen IT, Kolonay JF, Brinkac L, Beanan M, Dodson RJ, Daugherty SC, Madupu R, Angiuoli SV, Durkin AS, Haft DH, Vamathevan J, Khouri H, Utterback T, Lee C, Dimitrov G, Jiang L, Qin H, Weidman J, Tran K, Kang K, Hance IR, Nelson KE, Fraser CM. “Insights on Evolution of Virulence and Resistance from the Complete Genome Analysis of an Early Methicillin-Resistant Staphylococcus aureus Strain and a Biofilm-Producing Methicillin-Resistant Staphylococcus epidermidis Strain”. (2005) Journal of Bacteriology 187: 2426-2438.
- ↑ 11.0 11.1 11.2 11.3 11.4 11.5 11.6 Heilmann C., Hussain M., Peters G., Götz F. "Evidence for autolysin-mediated primary attachment of Staphylococcus epidermidis to a polystyrene surface." (1997) Mol. Microbiol. 24: 1013–1024.
- ↑ 12.0 12.1 Bowden M. G., Visai L., Longshaw C. M., Holland K. T., Speziale P., Hook M. "Is the GehD lipase from Staphylococcus epidermidis a collagen binding adhesin?" (2002).J. Biol. Chem. 277: 43017–43023.
- ↑ McCrea K. W., Hartford O., Davis S., Eidhin D. N., Lina G., Speziale P., et al. "The serine-aspartate repeat (Sdr) protein family in Staphylococcus epidermidis." (2000) Microbiology 146: 1535–1546.
- ↑ Arrecubieta C., Asai T., Bayern M., Loughman A., Fitzgerald J. R., Shelton C. E., Baron, H.M., Dang, N.C., Deng, M.C., Naka,Y., Foster, T.J., Lowy, F.D. "The role of Staphylococcus aureus adhesins in the pathogenesis of ventricular assist device-related infections." (2006) J. Infect. Dis. 193:1109–1119.
- ↑ Costerton J. W., Lewandowski Z., Caldwell D. E., Korber D. R., Lappin-Scott H. M. "Microbial biofilms." (1995). Annu. Rev. Microbiol. 49: 711–745.
- ↑ Shahrooei M., Hira V., Stijlemans B., Merckx R., Hermans P. W., Van Eldere J. " Inhibition of Staphylococcus epidermidis biofilm formation by rabbit polyclonal antibodies against the SesC protein." (2009) Infect. Immun. 77: 3670–3678.
- ↑ Mack, D., Rohde, H., Dobinsky, S., Riedewald, J., Nedelmann, M., Knobloch, J., Elsner, H. and Hubert Feucht. "Identification of Three Essential Regulatory Gene Loci Governing Expression of Staphylococcus epidermidis Polysaccharide Intercellular Adhesin and Biofilm Formation" (2000) Infect. Immun.. 68: 3799-3807.
- ↑ 18.0 18.1 Gahlot, Rupam et al. “Catheter-Related Bloodstream Infections.” (2014) International Journal of Critical Illness and Injury Science 4.2: 162–167. PMC.
- ↑ Dadwal, S., Kriengkauykiat, J., and James Ito. "Infectious Complications." Infectious Complications. Cancer Network, 01 Nov. 2015. Web. 20 Apr. 2017.
- ↑ Raucher H S, Hyatt A C, Barzilai A, et al. “Quantitative blood cultures in the evaluation of septicemia in children with Broviac catheters.” (1984) J Pediatr. 104:29–33.
- ↑ “Broad Spectrum Antibiotics : Uses, Advantages & Disadvantages.” SRS PHARMACEUTICALS PVT. LTD, 1995. Web. 20 Apr. 2017.
- ↑ Bullard KM, Dunn DL. “Bloodstream and intravascular catheter infections.” In: Holzheimer RG, Mannick JA, editors. Surgical Treatment: Evidence-Based and Problem-Oriented. Munich: Zuckschwerdt; 2001
- ↑ 23.0 23.1 "Your Implanted Port." Memorial Sloan Kettering Cancer Center. 18 Aug. 2016. Web. 22 Apr. 2017.
Authored for BIOL 238 Microbiology, taught by Joan Slonczewski, 2017, Kenyon College.
