Acidithiobacillus albertensis: Difference between revisions
m (→Species) |
|||
| Line 19: | Line 19: | ||
Describe the size and content of the genome. How many chromosomes? Circular or linear? Other interesting features? What is known about its sequence? | Describe the size and content of the genome. How many chromosomes? Circular or linear? Other interesting features? What is known about its sequence? | ||
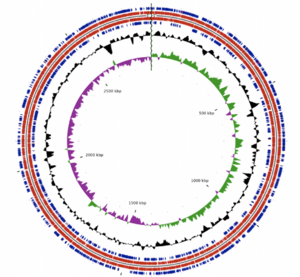
[[File:Albertensis_genome.png|thumb||right|The ''A. albertensis'' genome consists of a single, circular chromosome]] | |||
==Cell Structure, Metabolism and Life Cycle== | ==Cell Structure, Metabolism and Life Cycle== | ||
Revision as of 23:29, 24 April 2024
Classification
Domain; Bacteria Phylum; Proteobacteria Class; Acidithiobacillia Order; Acidithiobacillales family; Acidithiobacillaceae Genus: Acidithiobacillus
Species
|
NCBI: [1] |
Acidithiobacillus albertensis
Description and Significance
Describe the appearance, habitat, etc. of the organism, and why you think it is important.
Genome Structure
Describe the size and content of the genome. How many chromosomes? Circular or linear? Other interesting features? What is known about its sequence?
Cell Structure, Metabolism and Life Cycle
Interesting features of cell structure; how it gains energy; what important molecules it produces.
Ecology and Pathogenesis
Habitat; symbiosis; biogeochemical significance; contributions to environment.
If relevant, how does this organism cause disease? Human, animal, plant hosts? Virulence factors, as well as patient symptoms.
References
“Chalcopyrite.” Encyclopædia Britannica, Encyclopædia Britannica, inc., www.britannica.com/science/chalcopyrite.
Castro, M., et al. “Draft genome sequence of the type strain of the sulfur-oxidizing acidophile, Acidithiobacillus albertensis (DSM 14366)”. Standards in genomic sciences. 2017. Volume 12 p. 77-84.
Chen, J., Liu, Y., Diep, P., Mahadevan, R. “Genetic engineering of extremely acidophilic Acidithiobacillus species for biomining: Progress and perspectives”. Journal of Hazardous Materials. 2022. Volume 438 p. 129-247.
Moya-Beltrán, A., Beard, S., Rojas-Villalobos, C. et al. “Genomic evolution of the class Acidithiobacillia: deep-branching Proteobacteria living in extreme acidic conditions”. ISME J. 2021. Volume 15 p. 3221–3238.
Moya-Beltrán, A., Gajdosik, M., Rojas-Villalobos, C. et al. “Influence of mobile genetic elements and insertion sequences in long- and short-term adaptive processes of Acidithiobacillus ferrooxidans strains”. Sci Rep. 2023.Volume 13 p. 10876-10890.
Rzhepishevska, O. “Physiology and Genetics of Acidithiobacillus Species: Applications for Biomining”. Umeå University. 2023.
Schofield, E. “Acid Rock Drainage: More than Just a Mining Project Concern?” Barr Engineering Co., 2023. www.barr.com/Insights/Insights-Article/ArtMID/1344/ArticleID/380/Acid-rock-drainage-More-than-just-a-mining-project-concern.
“Section 3 - Acidithiobacillus: Safety Assessment of Transgenic Organisms”. OECD. 2006. Volume 2.
“Tetrathionate.” Wikipedia, Wikimedia Foundation, 27 Aug. 2023, en.wikipedia.org/wiki/Tetrathionate.
Vardanyan, N.S., and A.K. Vardanyan. “New sulphur oxidizing bacteria isolated from bioleaching pulp of zinc and copper concentrates.” Universal Journal of Microbiology Research. 2014. Volume 2 p. 27–31.
Xia, J., et al. “A new strain acidithiobacillus albertensis by-05 for bioleaching of metal sulfides ores.” Transactions of Nonferrous Metals Society of China. 2007. Volume 17 p. 168–175.
Author
Page authored by Ben Sauer and Rose Schnabel, students of Prof. Jay Lennon at IndianaUniversity.