Antibiotic Resistance Within Staphylococcus Aureus: Difference between revisions
No edit summary |
|||
| (37 intermediate revisions by the same user not shown) | |||
| Line 10: | Line 10: | ||
[[File:Staphylococcus aureus VISA 2.jpg|thumb|[https://commons.wikimedia.org/wiki/Staphylococcus_aureus#/media/File:Staphylococcus_aureus_VISA_2.jpg Figure 2]. 2000x magnified image of methicillin resistant <i>S. aureus</i> bacteria under high magnification]] | [[File:Staphylococcus aureus VISA 2.jpg|thumb|[https://commons.wikimedia.org/wiki/Staphylococcus_aureus#/media/File:Staphylococcus_aureus_VISA_2.jpg Figure 2]. 2000x magnified image of methicillin resistant <i>S. aureus</i> bacteria under high magnification]] | ||
<i>Staphylococcus aureus</i> was first discovered by Alexander Ogston in 1880 when he became interested in the high post surgery mortality rates. The bacteria was later named by Frederich Julius Rosenbach. More commonly referred to as a “staph infection” or “staph bacteria”, <i>S. aureus</i> is reportedly found in 30% of the population who are asymptomatic nasal carriers, while another large portion carry the bacteria on their skin <sup>[5]</sup>. Newborn babies have the highest rates of nasal staphylococci while the second largest carrier group are hospital personnel<sup>[6]</sup>. <i>S. aureus</i> is transferred through skin to skin contact and more commonly affects persons previously diseased or with weaker immune systems. The most common transfer happens from a health worker who has been in contact with an active strain of it. Other transfers occur from environmental sources or from other carriers of the bacteria. Possible outcomes from staph infections include many hospital originated issues including pneumonia and bloodstream and wound infections. When <i>S. aureus</i> was first identified, and before antibiotics were developed, the bacteria had a mortality rate of 80%. Today, with antibiotic treatment, the mortality rates around the world range from 20-40%, but with antibiotic resistance, the mortality rate is slowly increasing <sup>[7]</sup>. | <i>Staphylococcus aureus</i> was first discovered by Alexander Ogston in 1880 when he became interested in the high post surgery mortality rates. The bacteria was later named by Frederich Julius Rosenbach. More commonly referred to as a “staph infection” or “staph bacteria”, <i>S. aureus</i> is reportedly found in 30% of the population who are asymptomatic nasal carriers, while another large portion carry the bacteria on their skin <sup>[5]</sup>. Newborn babies have the highest rates of nasal staphylococci while the second largest carrier group are hospital personnel<sup>[6]</sup>. <i>S. aureus</i> is transferred through skin to skin contact and more commonly affects persons previously diseased or with weaker immune systems. The most common transfer happens from a health worker who has been in contact with an active strain of it. Other transfers occur from environmental sources or from other carriers of the bacteria. Possible outcomes from staph infections include many hospital originated issues including pneumonia and bloodstream and wound infections. When <i>S. aureus</i> was first identified, and before antibiotics were developed, the bacteria had a mortality rate of 80%. Today, with antibiotic treatment, the mortality rates around the world range from 20-40%, but with antibiotic resistance, the mortality rate is slowly increasing <sup>[7]</sup>. | ||
[[File:b lactam antibiotic overview.png|thumb|left|[http://commons.wikimedia.org/wiki/File:Penicillin_inhibition.svg#/media/File:Penicillin_inhibition.svg Figure 3]. Cell wall formation of <i>S. aureus</i> being inhibited by a β-lactam antibiotic through a binding protein.]] | |||
===Cell Structure=== | ===Cell Structure=== | ||
Bacteria interactions with antibiotics are based on the structure of the cell itself. <i>Staphylococcus aureus</i> is a gram positive bacteria that belongs to the [http://en.wikipedia.org/wiki/Micrococcaceae micrococcaceae] family and appears in the form of cocci clusters, as seen in Figure 2. It differs from other staphylococcus because it appears with gold pigmentation. The bacteria also tests positive for [http://en.wikipedia.org/wiki/Coagulase coagulase], [http://en.wikipedia.org/wiki/Mannitol_salt_agar mannitol-fermentation], and [http://en.wikipedia.org/wiki/Deoxyribonuclease deoxyribonuclease]. <i>Staphylococcus aureus</i> has a thick cell wall made up of peptidoglycan and in this case are 50 percent by weight. The peptidoglycan are made up of both N-acetylglucosamine and N-acetylmuramic acid, which are linked with [http://en.wikipedia.org/wiki/Tetrapeptide tetrapeptide chains]. Through [http://en.wikipedia.org/wiki/Morphology_(biology) morphological examination]the cell wall was found to contain peptidoglycan along with techoic acids and proteins <sup>[8]</sup>. [http://en.wikipedia.org/wiki/Teichoic_acid Techoic acids] are bound to the peptidoglycan through [http://en.wikipedia.org/wiki/Phosphodiester_bond phosphodiester bonds]. | |||
Bacteria interactions with antibiotics are based on the structure of the cell itself. <i>Staphylococcus aureus</i> is a gram positive bacteria that belongs to the [http://en.wikipedia.org/wiki/Micrococcaceae micrococcaceae] family and appears in the form of cocci clusters, as seen in Figure 2. It differs from other staphylococcus because it appears with gold pigmentation. The bacteria also tests positive for [http://en.wikipedia.org/wiki/Coagulase coagulase], [http://en.wikipedia.org/wiki/Mannitol_salt_agar mannitol-fermentation], and [http://en.wikipedia.org/wiki/Deoxyribonuclease deoxyribonuclease]. <i>Staphylococcus aureus</i> has a thick cell wall made up of peptidoglycan and in this case are 50 percent by weight. The peptidoglycan are made up of both N-acetylglucosamine and N-acetylmuramic acid, which are linked with [http://en.wikipedia.org/wiki/Tetrapeptide tetrapeptide chains]. Through [http://en.wikipedia.org/wiki/Morphology_(biology) morphological examination]the cell wall was found to contain peptidoglycan along with techoic acids and proteins <sup>[8]</sup>. [http://en.wikipedia.org/wiki/Teichoic_acid Techoic acids] are bound to the peptidoglycan through [http://en.wikipedia.org/wiki/Phosphodiester_bond phosphodiester bonds]. The acids then extend out where proteins are bound and form the outer most layer of the cell. Antibiotics work by binding to the proteins and inhibiting cell wall formation by inhibiting the cell enzyme which can be seen in Fig. 3. In this case, <i>Staphylococcus aureus</i> contains β-lactamase enzyme and β-lactam antibiotics are able to bind to this enzyme. The cell itself contains [http://en.wikipedia.org/wiki/Prophage prophages], [http://en.wikipedia.org/wiki/Plasmid plasmids], and [http://en.wikipedia.org/wiki/Transposable_element transposons] all within a 2800 bp circular chromosome <sup>[5]</sup>. This is where mechanisms of antibiotic resistance are found and transferred between cells through horizontal gene transfer. | The acids then extend out where proteins are bound and form the outer most layer of the cell. Antibiotics work by binding to the proteins and inhibiting cell wall formation by inhibiting the cell enzyme which can be seen in Fig. 3. In this case, <i>Staphylococcus aureus</i> contains β-lactamase enzyme and β-lactam antibiotics are able to bind to this enzyme. The cell itself contains [http://en.wikipedia.org/wiki/Prophage prophages], [http://en.wikipedia.org/wiki/Plasmid plasmids], and [http://en.wikipedia.org/wiki/Transposable_element transposons] all within a 2800 bp circular chromosome <sup>[5]</sup>. This is where mechanisms of antibiotic resistance are found and transferred between cells through horizontal gene transfer. | ||
==Penicillin Resistance: History and Mechanisms== | ==Penicillin Resistance: History and Mechanisms== | ||
| Line 23: | Line 25: | ||
===Resistance=== | ===Resistance=== | ||
[[File:penicillin b ring.gif|thumb|[http://www.wiley.com/college/pratt/0471393878/instructor/activities/bacterial_drug_resistance/index.html Figure 4]. β-lactam ring is hydrolyzed by bacteria.]] | |||
β-lactam antibiotics, like penicillin, work by inhibiting the cell enzyme which in this case would be β-lactamase <sup>[4]</sup>. Inhibiting the cell enzyme slows down cell wall formation which brings down osmotic pressure and eventually kills the cell. β-lactam antibiotics are able to inhibit enzymes with a [http://en.wikipedia.org/wiki/Β-Lactam β-lactam ring], which binds to the enzyme in the bacteria cell. One way in which bacteria become resistant to antibiotics is by hydrolyzing the β-lactam ring (shown in Fig. 4). They do this when they contain a [https://www.wikigenes.org/e/gene/e/2598287.html blaZ gene], which codes for penicillinase. The gene reorganizes the enzyme and does not allow for it to bind with β-lactam ring. BlaZ gene is located on a transposable part of the large plasmid within the <i>S. aureus </i> bacteria cells. Because of this location, the gene is easily movable to surrounding cells through [http://en.wikipedia.org/wiki/Horizontal_gene_transfer horizontal gene transfer]. | β-lactam antibiotics, like penicillin, work by inhibiting the cell enzyme which in this case would be β-lactamase <sup>[4]</sup>. Inhibiting the cell enzyme slows down cell wall formation which brings down osmotic pressure and eventually kills the cell. β-lactam antibiotics are able to inhibit enzymes with a [http://en.wikipedia.org/wiki/Β-Lactam β-lactam ring], which binds to the enzyme in the bacteria cell. One way in which bacteria become resistant to antibiotics is by hydrolyzing the β-lactam ring (shown in Fig. 4). They do this when they contain a [https://www.wikigenes.org/e/gene/e/2598287.html blaZ gene], which codes for penicillinase. The gene reorganizes the enzyme and does not allow for it to bind with β-lactam ring. BlaZ gene is located on a transposable part of the large plasmid within the <i>S. aureus </i> bacteria cells. Because of this location, the gene is easily movable to surrounding cells through [http://en.wikipedia.org/wiki/Horizontal_gene_transfer horizontal gene transfer]. | ||
| Line 32: | Line 35: | ||
===Resistance=== | ===Resistance=== | ||
[[File:mecA structrue.jpg|thumb|[http://www.thelancet.com/journals/laninf/article/PIIS1473-3099(11)70126-8/fulltext?version=printerFriendly Figure 5]. MecA gene structure for methicillin resistant <i>S. aureus</i> bacteria (top). CcrA and CcrB are both present.]] | |||
The mecA gene is located within the chromosome which allows the bacteria cell to transfer it to other cells. In a study done by Katayama et all, it was found that all methicillin resistant strands of <i>S. aureus</i> contain the mecA gene, confirming previous studies that identified this as the resistant coding gene. The mecA gene is regulated by two [http://en.wikipedia.org/wiki/Recombinase recombinase genes] including the ccrA and the ccrB. The structure of the mecA gene can be seen in Figure 5. Through [http://en.wikipedia.org/wiki/Polymerase_chain_reaction PCR], DNA was extracted in order to remove the mecA gene. In this process, it was proven that the excision of SCCmec DNAs caused a loss of resistance in <i>S. aureus</i>. Taking this further, Katayama et all investigated the role of plasmids in the transfer of resistance mechanisms. Through [http://en.wikipedia.org/wiki/Electroporation electroporation], recombinant plasmids were introduced into a wild type strain and cultured on an agar plate containing methicillin. The bacteria, <i>S. aureus</i>, still grew multiple colonies illustrating that the mecA gene could only be expressed if ccrA and ccrB are present in the plasmids <sup>[10]</sup>. To summarize, based on this study and previous ones, methicillin resistance within <i>Staphylococcus aureus</i> only works when the MecA gene is present along with ccrA and ccrB which help express the MecA gene. The gene inhibits protein production and makes antibiotic binding more difficult. If MecA gene is present but without ccrA or ccrB, the gene still produces proteins, but those proteins are structured to bind with β-lactam rings that are found in methicillin. | The mecA gene is located within the chromosome which allows the bacteria cell to transfer it to other cells. In a study done by Katayama et all, it was found that all methicillin resistant strands of <i>S. aureus</i> contain the mecA gene, confirming previous studies that identified this as the resistant coding gene. The mecA gene is regulated by two [http://en.wikipedia.org/wiki/Recombinase recombinase genes] including the ccrA and the ccrB. The structure of the mecA gene can be seen in Figure 5. Through [http://en.wikipedia.org/wiki/Polymerase_chain_reaction PCR], DNA was extracted in order to remove the mecA gene. In this process, it was proven that the excision of SCCmec DNAs caused a loss of resistance in <i>S. aureus</i>. Taking this further, Katayama et all investigated the role of plasmids in the transfer of resistance mechanisms. Through [http://en.wikipedia.org/wiki/Electroporation electroporation], recombinant plasmids were introduced into a wild type strain and cultured on an agar plate containing methicillin. The bacteria, <i>S. aureus</i>, still grew multiple colonies illustrating that the mecA gene could only be expressed if ccrA and ccrB are present in the plasmids <sup>[10]</sup>. To summarize, based on this study and previous ones, methicillin resistance within <i>Staphylococcus aureus</i> only works when the MecA gene is present along with ccrA and ccrB which help express the MecA gene. The gene inhibits protein production and makes antibiotic binding more difficult. If MecA gene is present but without ccrA or ccrB, the gene still produces proteins, but those proteins are structured to bind with β-lactam rings that are found in methicillin. | ||
| Line 37: | Line 41: | ||
==Fighting Antibiotic Resistance in Staphylococcus Aureus Today== | ==Fighting Antibiotic Resistance in Staphylococcus Aureus Today== | ||
As <i>S. aureus</i> becomes resistant to more and more antibitoics, many attempts have been made to find other ways to treat infections. Most recently, it was discovered that a [http://www.bbc.com/news/uk-england-nottinghamshire-32117815 medieval remedy of garlic and part of a cow’s stomach] could kill the superbug. No official research on this remedy has been published yet. Another attempt made has been through [http://en.wikipedia.org/wiki/Phage_therapy phage therapy]. In a study performed by Capparelli et all in 2007, phage treatment was found successful in treating and saving 97% of mice that were injected with the <i>S. aureus</i> bacteria and a phage. The <i>S. aureus</i> bacteria strains were taken directly from patients in the hospital who were infected with multi-antibiotic resistant strains, grown on agar plates, and eventually confirmed as <i>S. aureus</i> bacteria through [http://en.wikipedia.org/wiki/Coagulase#Coagulase_test coagulase tests], microscopic observation, and PCR. For the bacteriophages, twenty different strains of <i>S. aureus</i> bacteria were grown in [http://en.wikipedia.org/wiki/Lysogeny_broth LB broth] and [http://en.wikipedia.org/wiki/Mitomycin mitomycin C] was added. Supernatants were screened for phages and the phage DNA was tested and found to contain genes eta, tst, lukSPV-lukFPV, and sea. The phage that proved to have the largest host range contained only lukSPV-lukFPV, and sea genes and was called the wild phage. This phage was injected into mice with one strain of the bacteria which allowed for the isolation of another phage which can be referred to as phage M. Both phages were tested in mice and wild phage was found to kill <i>S. aureus</i>, but was also found to cause harmful effects in both the liver and spleen after 2 days. Phage M continued to fight bacteria in the mice for 20-25 days. This difference can be attributed to the difference in antigens presented on the surface of each bacteriophage. Phage M has less antigens and therefore antibodies used to fight it are more specific and target the phage more directly. Phage W allowed for more antibodies to target other bacteria in a less specific way causing harm to the body. Phage M was found to successfully kill <i>S. aureus</i> bacteria both [http://en.wikipedia.org/wiki/ | As <i>S. aureus</i> becomes resistant to more and more antibitoics, many attempts have been made to find other ways to treat infections. Most recently, it was discovered that a [http://www.bbc.com/news/uk-england-nottinghamshire-32117815 medieval remedy of garlic and part of a cow’s stomach] could kill the superbug. No official research on this remedy has been published yet. Another attempt made has been through [http://en.wikipedia.org/wiki/Phage_therapy phage therapy]. In a study performed by Capparelli et all in 2007, phage treatment was found successful in treating and saving 97% of mice that were injected with the <i>S. aureus</i> bacteria and a phage. [[File:in vitro.gif|thumb|left|Figure 6.Number of live bacteria treated and untreated with M phage. (A) Amount of live bacteria recovered after being preincubated with M phage was found to be much lower than untreated (⧫).[http://aac.asm.org/content/51/8/2765.full Capparelli et all, 2007]]] | ||
[[File:in vivo.gif|thumb|right|Figure 7.Number of mice surviving after being injected with <i>S. aureus</i> (A) and number of mice surviving after being injected with both <i>S. aureus</i> and M phage (B). [http://aac.asm.org/content/51/8/2765.full Capparelli et all, 2007]]] | |||
The <i>S. aureus</i> bacteria strains were taken directly from patients in the hospital who were infected with multi-antibiotic resistant strains, grown on agar plates, and eventually confirmed as <i>S. aureus</i> bacteria through [http://en.wikipedia.org/wiki/Coagulase#Coagulase_test coagulase tests], microscopic observation, and PCR. For the bacteriophages, twenty different strains of <i>S. aureus</i> bacteria were grown in [http://en.wikipedia.org/wiki/Lysogeny_broth LB broth] and [http://en.wikipedia.org/wiki/Mitomycin mitomycin C] was added. Supernatants were screened for phages and the phage DNA was tested and found to contain genes eta, tst, lukSPV-lukFPV, and sea. The phage that proved to have the largest host range contained only lukSPV-lukFPV, and sea genes and was called the wild phage. This phage was injected into mice with one strain of the bacteria which allowed for the isolation of another phage which can be referred to as phage M. Both phages were tested in mice and wild phage was found to kill <i>S. aureus</i>, but was also found to cause harmful effects in both the liver and spleen after 2 days. Phage M continued to fight bacteria in the mice for 20-25 days. This difference can be attributed to the difference in antigens presented on the surface of each bacteriophage. Phage M has less antigens and therefore antibodies used to fight it are more specific and target the phage more directly. Phage W allowed for more antibodies to target other bacteria in a less specific way causing harm to the body. Phage M was found to successfully kill <i>S. aureus</i> bacteria both [http://en.wikipedia.org/wiki/In_vitro in vitro] and [http://en.wikipedia.org/wiki/In_vivo in vivo], which can be seen in Figures 6 and 7, respectively. As is illustrated in this paper, more methods are being developed to combat antibiotic resistance, but no way is a “cure all” because bacteria continues to develop resistance mechanisms <sup>[11]</sup>. | |||
==References== | ==References== | ||
<sup>[1]</sup> [http://www.cdc.gov/drugresistance/threat-report-2013/ "Antibiotic Resistance Threats in the United States, 2013." Centers for Disease Control and Prevention. Centers for Disease Control and Prevention, 17 July 2014. Web. 9 Apr. 2015.] | |||
<sup>[2]</sup> [http://www.annualreviews.org/doi/pdf/10.1146/annurev.bi.42.070173.002351 Benveniste, Raoul and Davies, Julian. “Mechanisms of Antibiotic Resistance in Bacteria”. nnu. Rev. Biochem. (1973): 42:471-506. Print.] | <sup>[2]</sup> [http://www.annualreviews.org/doi/pdf/10.1146/annurev.bi.42.070173.002351 Benveniste, Raoul and Davies, Julian. “Mechanisms of Antibiotic Resistance in Bacteria”. nnu. Rev. Biochem. (1973): 42:471-506. Print.] | ||
<sup>[3]</sup>[http://wwwnc.cdc.gov/eid/article/13/12/07-0629_article "Hospitalizations and Deaths Caused by Methicillin-Resistant Staphylococcus Aureus, United States, 1999–2005 - Volume 13, Number 12-December 2007 - Emerging Infectious Disease Journal - CDC." Hospitalizations and Deaths Caused by Methicillin-Resistant Staphylococcus Aureus, United States, 1999–2005 - Volume 13, Number 12-December 2007 - Emerging Infectious Disease Journal - CDC. N.p., n.d. Web. 9 Apr. 2015] | |||
<sup>[4]</sup> [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC154455/ Lowy, Franklin D. “Antimicrobial Resistance: The Example of Staphylococcus Aureus.” Journal of Clinical Investigation 111.9 (2003): 1265–1273. PMC. Web. 15 Feb. 2015] | <sup>[4]</sup> [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC154455/ Lowy, Franklin D. “Antimicrobial Resistance: The Example of Staphylococcus Aureus.” Journal of Clinical Investigation 111.9 (2003): 1265–1273. PMC. Web. 15 Feb. 2015] | ||
<sup>[5]</sup> [http://www.nejm.org/doi/full/10.1056/NEJM199808203390806 Lowy, Franklin D. "Staphylococcus Aureus Infections." N Engl J Med 339.8 (1998): 520-32. Print] | <sup>[5]</sup> [http://www.nejm.org/doi/full/10.1056/NEJM199808203390806 Lowy, Franklin D. "Staphylococcus Aureus Infections." N Engl J Med 339.8 (1998): 520-32. Print] | ||
<sup>[6]</sup> [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC441169/pdf/bactrev00136-0060.pdf Williams, R. E. O. “HEALTHY CARRIAGE OF STAPHYLOCOCCUS AUREUS: ITS PREVALENCE AND IMPORTANCE.” Bacteriological Reviews 27.1 (1963): 56–71. Print.] | |||
<sup>[7]</sup> [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC154455/ Lowy, Franklin D. “Antimicrobial Resistance: The Example of Staphylococcus Aureus.” Journal of Clinical Investigation 111.9 (2003): 1265–1273. PMC. Web. 15 Feb. 2015.] | <sup>[7]</sup> [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC154455/ Lowy, Franklin D. “Antimicrobial Resistance: The Example of Staphylococcus Aureus.” Journal of Clinical Investigation 111.9 (2003): 1265–1273. PMC. Web. 15 Feb. 2015.] | ||
| Line 66: | Line 68: | ||
<sup>[11]</sup> [http://aac.asm.org/content/51/8/2765.full Capparelli, Rosanna. Parlato, Marianna. Borriello, Giorgia. Salvatore, Paola. And Inannelli, Domenic. “Experimental Phage Therapy against <i>Staphylococcus Aureus</i> in Mice.” American Society for Microbiology. 51.8. (2007). Print.] | <sup>[11]</sup> [http://aac.asm.org/content/51/8/2765.full Capparelli, Rosanna. Parlato, Marianna. Borriello, Giorgia. Salvatore, Paola. And Inannelli, Domenic. “Experimental Phage Therapy against <i>Staphylococcus Aureus</i> in Mice.” American Society for Microbiology. 51.8. (2007). Print.] | ||
<!--Do not remove this line--> | |||
Edited by (Liza Ach), a student of [http://www.jsd.claremont.edu/faculty/profile.asp?FacultyID=254 Nora Sullivan] in BIOL168L (Microbiology) in [http://www.jsd.claremont.edu/ The Keck Science Department of the Claremont Colleges] Spring 2014. | |||
<!--Do not edit or remove this line-->[[Category:Pages edited by students of Nora Sullivan at the Claremont Colleges]] | |||
Latest revision as of 22:15, 28 April 2015

Antibiotic resistance is a growing world-wide issue because of its effect on the rapid spread of threatening diseases and infections and the inability to control them. Resistance occurs both naturally and due to human action. While natural resistance is a large issue, recently the involvement of modern medicine and overuse of antibiotics have caused about 23,000 people to die each year due to antibiotic resistant bacterial infections, while about 2 million people experience these same infections [1]. Antibiotic resistant infections can occur anywhere in the community but recently there has been an increase of infections that come from hospitals and other medical facilities. Resistance in bacteria is a large issue because as bacteria evolves and forms ways of counteracting the antibiotics, the antibiotics become ineffective and harmful and fatal bacteria is able to thrive in multiple environments. Resistance mechanisms can be divided into two categories; (1) chemical interaction that affects the binding between a target site and the antibiotic and (2) antibiotic inactivation [2]. Antibiotic inactivation mechanisms are more commonly associated with bacterium that gain resistance overtime from human action. An example of this can be traced through Staphylococcus aureus, which fights antibiotics by deactivating β-lactam binding proteins. Staphylococcus aureus is a gram positive bacteria that is present in the human microbiota, as well as the community and hospital environments. The presence of S. aureus on the skin microbiota can actually be beneficial to humans because it enables and expands the memory of T-cells, but if the bacteria infects the body, it becomes extremely dangerous and in some cases fatal. In 2005, more than 477,000 people were hospitalized due to S. aureus infections [3], and from that the mortality rate is about 20-40% [4].When the bacteria was first discovered in 1880, it was easily fought with penicillin and eventually methicillin. Today, S. aureus is a growing issue both within hospitals and communities because of its internal virulence and ability to adjust to different environments. The bacteria remains a growing pandemic, as shown in Fig. 1, because of its increasing antibiotic resistance to penicillin and methicillin through mechanisms including horizontal gene transfer and altering of antibiotics.
Staphylococcus Aureus
History and Background Information

Staphylococcus aureus was first discovered by Alexander Ogston in 1880 when he became interested in the high post surgery mortality rates. The bacteria was later named by Frederich Julius Rosenbach. More commonly referred to as a “staph infection” or “staph bacteria”, S. aureus is reportedly found in 30% of the population who are asymptomatic nasal carriers, while another large portion carry the bacteria on their skin [5]. Newborn babies have the highest rates of nasal staphylococci while the second largest carrier group are hospital personnel[6]. S. aureus is transferred through skin to skin contact and more commonly affects persons previously diseased or with weaker immune systems. The most common transfer happens from a health worker who has been in contact with an active strain of it. Other transfers occur from environmental sources or from other carriers of the bacteria. Possible outcomes from staph infections include many hospital originated issues including pneumonia and bloodstream and wound infections. When S. aureus was first identified, and before antibiotics were developed, the bacteria had a mortality rate of 80%. Today, with antibiotic treatment, the mortality rates around the world range from 20-40%, but with antibiotic resistance, the mortality rate is slowly increasing [7].

Cell Structure
Bacteria interactions with antibiotics are based on the structure of the cell itself. Staphylococcus aureus is a gram positive bacteria that belongs to the micrococcaceae family and appears in the form of cocci clusters, as seen in Figure 2. It differs from other staphylococcus because it appears with gold pigmentation. The bacteria also tests positive for coagulase, mannitol-fermentation, and deoxyribonuclease. Staphylococcus aureus has a thick cell wall made up of peptidoglycan and in this case are 50 percent by weight. The peptidoglycan are made up of both N-acetylglucosamine and N-acetylmuramic acid, which are linked with tetrapeptide chains. Through morphological examinationthe cell wall was found to contain peptidoglycan along with techoic acids and proteins [8]. Techoic acids are bound to the peptidoglycan through phosphodiester bonds. The acids then extend out where proteins are bound and form the outer most layer of the cell. Antibiotics work by binding to the proteins and inhibiting cell wall formation by inhibiting the cell enzyme which can be seen in Fig. 3. In this case, Staphylococcus aureus contains β-lactamase enzyme and β-lactam antibiotics are able to bind to this enzyme. The cell itself contains prophages, plasmids, and transposons all within a 2800 bp circular chromosome [5]. This is where mechanisms of antibiotic resistance are found and transferred between cells through horizontal gene transfer.
Penicillin Resistance: History and Mechanisms
History and Background
The first antibiotic introduced to fight S. aureus was penicillin in 1940, which decreased mortality rates significantly. Two years later, resistance to the drug was already being recognized both within communities and hospitals. More than 80% of staphylococcus strains were found to be resistant by the early 1960’s. These strains were found by Bondi and Dietz in 1948 to contain penicillinase, which is a specific type of β-lactamase.
Resistance

β-lactam antibiotics, like penicillin, work by inhibiting the cell enzyme which in this case would be β-lactamase [4]. Inhibiting the cell enzyme slows down cell wall formation which brings down osmotic pressure and eventually kills the cell. β-lactam antibiotics are able to inhibit enzymes with a β-lactam ring, which binds to the enzyme in the bacteria cell. One way in which bacteria become resistant to antibiotics is by hydrolyzing the β-lactam ring (shown in Fig. 4). They do this when they contain a blaZ gene, which codes for penicillinase. The gene reorganizes the enzyme and does not allow for it to bind with β-lactam ring. BlaZ gene is located on a transposable part of the large plasmid within the S. aureus bacteria cells. Because of this location, the gene is easily movable to surrounding cells through horizontal gene transfer.
Methicillin: History and Resistance
History and Background
In 1961, methicillin was introduced as an attempt to combat pencillin resistant staphylococcus aureus strains. In less than a year, some S. aureus were reported to be methicillin resistant. Beginning in hospitals in the 1970s, and slowly moving out to the community, methicillin resistant S. aureus (MRSA) became a growing issue. Similar to penicillin, methicillin works by inhibiting the synthesis of cell wall through stoppage of peptidoglycan formation. Peptidoglycan for S. aureus is, as mentioned earlier, externally bound to penicillin binding proteins. These proteins both cross link and extend the peptidoglycan chain that form the cell wall. Methicillin works by inhibiting these proteins, thus stopping cell wall formation and making the cell weak and eventually die. Over time, S. aureus has built resistance to this type of antibiotic. This is attributed to the mecA gene, which is actually transferred from different bacteria and can be traced back to Staphylococcus sciuri [9]. When the gene is present, penicillin binding proteins are still produced, but their structure make it harder for β-lactam rings within the antibiotics to bind and stop cell wall formation. This mechanism is similar to that of penicillin and the blaZ gene.
Resistance
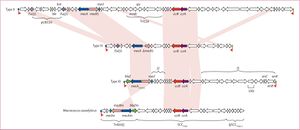
The mecA gene is located within the chromosome which allows the bacteria cell to transfer it to other cells. In a study done by Katayama et all, it was found that all methicillin resistant strands of S. aureus contain the mecA gene, confirming previous studies that identified this as the resistant coding gene. The mecA gene is regulated by two recombinase genes including the ccrA and the ccrB. The structure of the mecA gene can be seen in Figure 5. Through PCR, DNA was extracted in order to remove the mecA gene. In this process, it was proven that the excision of SCCmec DNAs caused a loss of resistance in S. aureus. Taking this further, Katayama et all investigated the role of plasmids in the transfer of resistance mechanisms. Through electroporation, recombinant plasmids were introduced into a wild type strain and cultured on an agar plate containing methicillin. The bacteria, S. aureus, still grew multiple colonies illustrating that the mecA gene could only be expressed if ccrA and ccrB are present in the plasmids [10]. To summarize, based on this study and previous ones, methicillin resistance within Staphylococcus aureus only works when the MecA gene is present along with ccrA and ccrB which help express the MecA gene. The gene inhibits protein production and makes antibiotic binding more difficult. If MecA gene is present but without ccrA or ccrB, the gene still produces proteins, but those proteins are structured to bind with β-lactam rings that are found in methicillin.
Fighting Antibiotic Resistance in Staphylococcus Aureus Today
As S. aureus becomes resistant to more and more antibitoics, many attempts have been made to find other ways to treat infections. Most recently, it was discovered that a medieval remedy of garlic and part of a cow’s stomach could kill the superbug. No official research on this remedy has been published yet. Another attempt made has been through phage therapy. In a study performed by Capparelli et all in 2007, phage treatment was found successful in treating and saving 97% of mice that were injected with the S. aureus bacteria and a phage.
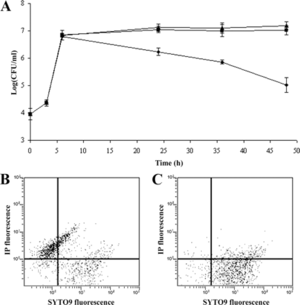
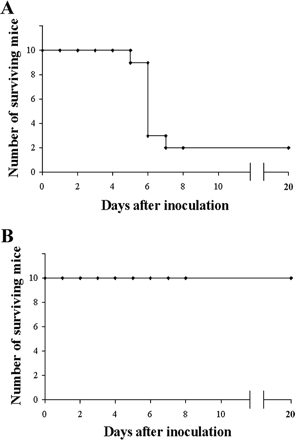
The S. aureus bacteria strains were taken directly from patients in the hospital who were infected with multi-antibiotic resistant strains, grown on agar plates, and eventually confirmed as S. aureus bacteria through coagulase tests, microscopic observation, and PCR. For the bacteriophages, twenty different strains of S. aureus bacteria were grown in LB broth and mitomycin C was added. Supernatants were screened for phages and the phage DNA was tested and found to contain genes eta, tst, lukSPV-lukFPV, and sea. The phage that proved to have the largest host range contained only lukSPV-lukFPV, and sea genes and was called the wild phage. This phage was injected into mice with one strain of the bacteria which allowed for the isolation of another phage which can be referred to as phage M. Both phages were tested in mice and wild phage was found to kill S. aureus, but was also found to cause harmful effects in both the liver and spleen after 2 days. Phage M continued to fight bacteria in the mice for 20-25 days. This difference can be attributed to the difference in antigens presented on the surface of each bacteriophage. Phage M has less antigens and therefore antibodies used to fight it are more specific and target the phage more directly. Phage W allowed for more antibodies to target other bacteria in a less specific way causing harm to the body. Phage M was found to successfully kill S. aureus bacteria both in vitro and in vivo, which can be seen in Figures 6 and 7, respectively. As is illustrated in this paper, more methods are being developed to combat antibiotic resistance, but no way is a “cure all” because bacteria continues to develop resistance mechanisms [11].
References
[5] Lowy, Franklin D. "Staphylococcus Aureus Infections." N Engl J Med 339.8 (1998): 520-32. Print
Edited by (Liza Ach), a student of Nora Sullivan in BIOL168L (Microbiology) in The Keck Science Department of the Claremont Colleges Spring 2014.
