EHEC
Etiology/Bacteriology
Taxonomy
| Domain = Bacteria | Phylum = Proteobacteria | Class = Gammaproteobacteria | Order = Enterobacteriales | Family = Enterobacteriaceae | Genus = Escherichia | species = E. coli | subspecies = O157:H7 | prototype strain = EDL933 |
|
NCBI: Taxonomy Genome: Escherichia coli O157:H7 str. EDL933 |
Description
Escherichia coli O157:H7 is an emerging pathogen that causes hemorrhagic colitis. E. coli is a Gram negative rod, a facultative anaerobe motile by means of peritrichous flagella, and found in the GI tract of mammals. Most E. coli are harmless and actually are an important members of a healthy human intestinal microbial community. However, E. coli is a very diverse group of bacteria consisting of many ecotypes (the harmless variety) and pathotypes. The pathogenic strains of E. coli can cause diarrheal diseases, urinary tract infections, and even meningitis. The E. coli pathotypes that cause diarrhea are transmitted via contaminated food or water, or through contact with infected animals or people. E. coli O157:H7 was first reported by the CDC in 1982. [1] E. coli O157:H7 is one of several pathotypes classified as enterohemorrhagic E. coli (EHEC). EHEC cause disease by producing a toxin called Shiga toxin, Shiga toxin causes bloody diarrhea and in approximately 5-10% of cases hemolytic uremic syndrome (HUS), which can lead to complete kidney failure. Patients with bloody diarrhea usually recover within a week and those who develop HUS usually recover within a few weeks, but in cases of kidney failure the patient may die. While people of any age are susceptible, the young and elderly are more often affected. Treatment of EHEC typically involves rehydration without administration of antibiotics and hospitalization in severe cases, especially HUS. EHEC infection is prevented by good hygiene, proper cooking of food, not swallowing water while swimming, and avoiding unpasteurized milk or apple cider. The prototypical E. coli O157:H7 strain is EDL933, which was isolated from Michigan ground beef that was linked to the original 1982 outbreak. The E. coli EDL933 genome was sequenced in 2001. [2] This strain of O157:H7 has been used in hundreds of scientific studies.
Pathogenesis
Transmission
E. coli O157:H7 was first isolated from hamburger and cattle are considered to be the primary reservoir for EHEC O157:H7. [3] Tissue tropism of EHEC at the rectal-anal junction and its stable colonization at this anatomical location ensures its persistence and shedding in feces. Not surprisingly, EHEC is often isolated from dairy farms and feedlots, including from house flies, although the latter is not necessarily a mechanical vector. Appearance in herds is seasonal, being highest during the summer, leading to the rancher's anecdote that the best preventative for EHEC is snow. Numerous recent EHEC outbreaks linked to lettuce and other leafy vegetables likely result from fecal contamination that occurs on farms and during production. The exceptionally tight adherence of E. coli O157:H7 to leaf surfaces and even within the stomata of spinach leaves no doubt contribute to its transmissibility. Essentially, any food source that is subject to fecal contamination can serve as a vehicle for food borne EHEC. This includes raw milk, unpasteurized apple cider, and even hazelnuts in shells, which suggests that EHEC is quite tolerant of drying.
Infectious dose, incubation, and colonization
Contributing substantially to transmission of EHEC is its extremely low infectious dose of as few as ten microorganisms. The low infectious dose also explains its spread from farm animals to humans and person-to-person transmission. The typical incubation period is 3 days, with a range of 1-8 days. E. coli O157:H7 readily colonizes the mammalian large intestine, including humans. In mice it is known that E. coli EDL933 colonizes the intestine by growing on sugars that are not used by at least some commensal E. coli ecotypes. Since a combination of 2 or more commensal ecotypes are able to prevent colonization of mice by E. coli O157:H7, this could explain attack rates in humans that vary from 2-50%, depending upon the nature of the colonized commensal ecotypes in the humans exposed to infection. In the more carefully documented outbreaks to date, which was associated with a contaminated rural water system in Alpine, WY, it was found that the attack rate was 23% for residents and 50% for visitors, which is consistent with partial immunity of the townspeople following long-term exposure. E. coli O157:H7 is not an invasive pathogen and causes disease by interaction with the intestinal mucosa. Furthermore, infected patients typically shed a billion bacteria per gram of diarrhaea. Hence, beginning with as few as ten infecting organisms, it is likely that E. coli O157:H7 must grow to populations exceeding a billion bacteria per gram of intestinal contents in order to cause disease.
Epidemiology
According to the CDC, the most prevalent EHEC transmission route since 1982 is foodborne, followed by person-to-person contact and waterborne infection. Following its emergence and initial outbreaks, the relative incidence of EHEC infections remains essentially unchanged. Interestingly, the relative number of EHEC outbreaks per year has increased since 1992, while the median size of the outbreaks has declined since 1990. Thus the overall rate of infection by EHEC in the US has stabilized, apparently because improved understanding of EHEC has resulted in diminished size of outbreaks despite the increased prevalence of outbreaks.
Virulence factors
EHEC virulence factors include the ability to adhere tightly to plant materials, acid tolerance, attachment and effacement of intestinal epithelium, and production of endotoxin and Shiga toxin. The regulator of "hyper-adherence", TdcR, and OmpA, an outer membrane protein that is expressed during hyper-adherence are implicated in binding of EHEC to alfalfa sprouts and seed coats. Loss of these virulence factors results in decreased adherence.
Survival of E. coli O157:H7 during passage through the stomach or encounter of acidic conditions in the environment is ensured by at least four distinct systems for acid tolerance. There are four corresponding acid resistance (AR) gene systems. The mechanism of AR1 is unknown. AR2, AR3, and AR4 each depend upon amino acid decarboxylation and consequent consumption of protons, whcih results in pH homeostasis. Expression of the AR systems is induced by acid environment, anaerobiosis, entry into stationary phase. Collectively, one or more of these systems is likely to be "on" when EHEC is exposed to acid, as would be expected to occur upon consumption by a potential host and subsequent passage through the stomach.
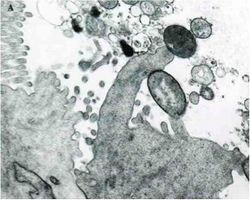
A cardinal virulence factor of EHEC is the Locus of Enterocyte Effacement (LEE), which is contained on a pathogenicity island that encodes all of the gene products needed for attaching and effacing the colonic epithelium. Following initial attachment of the EHEC cell to an epithelial cell, begins the second stage in which a number of LEE encoded proteins are secreted into the host cell. These proteins include EspA, EspB, and EspD. Translocation of these proteins into the host cell is essential for changing the host physiology. EspA may be part of the translocation machinery because it forms filamentous appendages between the bacterium and host cell. EspB is translocated and apparently alters host cell signal transduction pathways. These LEE proteins are secreted from EHEC by a type-III secretion system also encoded by LEE genes. A third stage of attachment and effacement involves intimate attachment of the EHEC cell to the host cell. To accomplish this, EHEC translocates its receptor protein, translocated intimin receptor (Tir), to the host cell where it is expressed on the host cell membrane. Simultaneously, EHEC expresses an a outer membrane protein called Intimin that is encoded by the eae gene. Thus EHEC encodes both its own receptor and its adhesin. Follwoing this tight associateion between the EHEC and host cells, actin pedestals are formed in the host cell, which push the host membrane to lengths up to 10 µm away from the host cell surface.
Clinical features
Diagnosis
Treatment
Prevention
Host Immune Response
References
1 Centers for Disease Control (CDC). Isolation of E. coli O157:H7 from sporadic cases of hemorrhagic colitis - United States. MMWR Morb Mortal Wkly Rep. 1982 Nov 5;31(43):580, 585. PubMed PMID: 6817062.
2 Perna NT, Plunkett G 3rd, Burland V, Mau B, Glasner JD, Rose DJ, Mayhew GF, Evans PS, Gregor J, Kirkpatrick HA, Pósfai G, Hackett J, Klink S, Boutin A, Shao Y, Miller L, Grotbeck EJ, Davis NW, Lim A, Dimalanta ET, Potamousis KD, Apodaca J, Anantharaman TS, Lin J, Yen G, Schwartz DC, Welch RA, Blattner FR. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature. 2001 Jan 25;409(6819):529-33. PubMed PMID: 11206551.
3 Hancock DD, Besser TE, Kinsel ML, Tarr PI, Rice DH, Paros MG. The prevalence of Escherichia coli O157.H7 in dairy and beef cattle in Washington State. Epidemiol Infect. 1994 Oct;113(2):199-207. PubMed PMID: 7925659.
4 Naylor SW, Low JC, Besser TE, Mahajan A, Gunn GJ, Pearce MC, McKendrick IJ, Smith DG, Gally DL. Lymphoid follicle-dense mucosa at the terminal rectum is the principal site of colonization of enterohemorrhagic Escherichia coli O157:H7 in the bovine host. Infect Immun. 2003 Mar;71(3):1505-12. PubMed PMID: 12595469.
Created by Tyrrell Conway at the University of Oklahoma.
