Extremophiles and Extraterrestrial Life: Difference between revisions
No edit summary |
|||
| Line 15: | Line 15: | ||
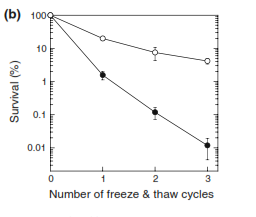
While Csps are present in some mesophiles, they are usually transiently expressed in response to cold shock, whereas in psychrophiles the proteins are constitutively expressed (as reviewed in Siddiqui et al. 2013). Most studies done on csps have been conducted on the mesophile Escherichia coli, an easier bacteria to cultivate than most psychrophiles. In a recent study examining the effects of psychrophile csps, E. coli genetically modified with a Psychromonas artica gene encoding for a cold shock protein showed a tenfold increase in cold tolerance (Jung et al. 2010). | While Csps are present in some mesophiles, they are usually transiently expressed in response to cold shock, whereas in psychrophiles the proteins are constitutively expressed (as reviewed in Siddiqui et al. 2013). Most studies done on csps have been conducted on the mesophile Escherichia coli, an easier bacteria to cultivate than most psychrophiles. In a recent study examining the effects of psychrophile csps, E. coli genetically modified with a Psychromonas artica gene encoding for a cold shock protein showed a tenfold increase in cold tolerance (Jung et al. 2010). | ||
[[Image:CSP_ecoli_Uli.png|thumb|300px|right| | [[Image:CSP_ecoli_Uli.png|thumb|300px|right|Survival rates of <i> Escherichia coli </i> expressing the control pAED4 gene (filled circle) and the <i> Psychromonas arctica </i> derived CspApa gene (open circle) after freeze/thaw cycles. <i> E. coli</i> expressing the psychrophile cold-shock protein genes had a 10x higher survival rate than control cells (Jung <i> et al </i>., 2010).]] | ||
Still, not all organisms have homologs of csps genes. Even organisms isolated from the same antartic lake can have differing csps genes, such as Methanogenium frigidum (does) and Methanococcoides burtonii (doesn't) (as reviewed in Siddiqui et al. 2013). M. burtonii, instead, upregulates Ctr (cold-responsive TRAM domain) proteins which might serve as RNA chaperones and respond to various stressful environments including increased methanol (as reviewed in Siddiqui et al. 2013). M. burtonii, an archaeon, expresses in lower temperatures a protein similar to DEAD-box RNA helicases (named for their Asp-Glu-Ala-Asp motif) found in some bacteria. The protein, which is predicted to have a postive charge, may work by attaching to the negative RNA backbone and then because of its central conserved domain bind to specific sequence targets on the RNA (Lim et al. 2000). One of the functions of DEAD box RNA helicases is to unwind RNA which has already formed the secondary structures (as reviewed in Siddiqui et al. 2013). | Still, not all organisms have homologs of csps genes. Even organisms isolated from the same antartic lake can have differing csps genes, such as Methanogenium frigidum (does) and Methanococcoides burtonii (doesn't) (as reviewed in Siddiqui et al. 2013). M. burtonii, instead, upregulates Ctr (cold-responsive TRAM domain) proteins which might serve as RNA chaperones and respond to various stressful environments including increased methanol (as reviewed in Siddiqui et al. 2013). M. burtonii, an archaeon, expresses in lower temperatures a protein similar to DEAD-box RNA helicases (named for their Asp-Glu-Ala-Asp motif) found in some bacteria. The protein, which is predicted to have a postive charge, may work by attaching to the negative RNA backbone and then because of its central conserved domain bind to specific sequence targets on the RNA (Lim et al. 2000). One of the functions of DEAD box RNA helicases is to unwind RNA which has already formed the secondary structures (as reviewed in Siddiqui et al. 2013). | ||
Revision as of 17:29, 26 May 2015
Introduction
Virtually everywhere on earth where water is present also has some sign of life. This is why water is such an important biosignature when found in extraterrestrial systems. Recent discovery of life on earth's most extreme environments has also made more possible the idea of panspermia and the transport of microbes within meteorites (Rothschild & Mancinelli 2001). This is especially relevant with the finding of polyextremophiles which can flourish under multiple physical (temperature, radiation) or geochemical extremes (dessication, pH, salinity) (Rothschild & Mancinelli 2001). Also important to study is the increased resistance among different stages of an organism's life cycle, such as spores, seeds, or egg stages (Rothschild & Mancinelli 2001).
Cold adaptation
It is important in the search for extraterrestial life to try to determine the upper and lower limits of growth and survival in order to better direct the searches. Since a majority of the solar system is made up of colder environments, it also makes sense to look into cold earth environments for organisms similar to those we might find (Reid et al. 2006). Conditions in the Vestfold Hills and the Dry Valleys in Antarctica, for example, can be models for some parts of Mars (Reid et al. 2006).
Cold adaptation can be achieved in a variety of ways, and has arisen in conjunction with adaptation to other environmental factors. This can be assumed due to the variety of environments that have extreme cold temperatures (such as deep sea, alpine regions, subterranean caverns, permafrost regions) and can present different in biotic and abiotic stresses on inhabiting organisms (Siddiqui et al. 2013). These circumstances have led to organisms capable of surviving in different extremes and good models for extraterrestrial life.
While their are many different survival mechanisms, most psychrophiles face similar stressors associated with cold temperatures which include decreased membrane fluidity, osmotic stress, and RNA folding (Siddiqui et al. 2013). Cold temperatures cause RNA to decrease in flexibility and form secondary transcript structures (Siddiqui et al. 2013). Organisms address this problem in a variety of ways, one of the most studied being the expression of cold shock proteins (Csps) which can attach to the RNA and keep secondary structures from forming.
While Csps are present in some mesophiles, they are usually transiently expressed in response to cold shock, whereas in psychrophiles the proteins are constitutively expressed (as reviewed in Siddiqui et al. 2013). Most studies done on csps have been conducted on the mesophile Escherichia coli, an easier bacteria to cultivate than most psychrophiles. In a recent study examining the effects of psychrophile csps, E. coli genetically modified with a Psychromonas artica gene encoding for a cold shock protein showed a tenfold increase in cold tolerance (Jung et al. 2010).
Still, not all organisms have homologs of csps genes. Even organisms isolated from the same antartic lake can have differing csps genes, such as Methanogenium frigidum (does) and Methanococcoides burtonii (doesn't) (as reviewed in Siddiqui et al. 2013). M. burtonii, instead, upregulates Ctr (cold-responsive TRAM domain) proteins which might serve as RNA chaperones and respond to various stressful environments including increased methanol (as reviewed in Siddiqui et al. 2013). M. burtonii, an archaeon, expresses in lower temperatures a protein similar to DEAD-box RNA helicases (named for their Asp-Glu-Ala-Asp motif) found in some bacteria. The protein, which is predicted to have a postive charge, may work by attaching to the negative RNA backbone and then because of its central conserved domain bind to specific sequence targets on the RNA (Lim et al. 2000). One of the functions of DEAD box RNA helicases is to unwind RNA which has already formed the secondary structures (as reviewed in Siddiqui et al. 2013).
Some archae and bacteria upregulate rnases and proteases to increase levels of synthetic precursors and remove damaged RNA and proteins (as reviewed in Siddiqui et al. 2013). Psychrobacter cryohalolentis has been shown to counteract the lowered catalytic and binding capacity of enzymes in cold temperatures by increasing the amount of ATP available in the cytoplasm, which is a substrate for many enzymatic reactions. More substrate, then, would help increase the rate of reactions in the cell. The increased levels of ATP at lower temperatures was not solely due to less usage of ATP, as increased levels of ADP were also present. Interestingly, cells showed a similar reaction to osmotic shock simulating the increased solute levels after ice formation (Amato & Christner 2008). Similarly, experiments done on a Escherichia coli mutant expressing higher ATP cytoplasm concentrations showed improved cold-tolerance at 0°C than a wild type (As cited in Amato, Morrison).
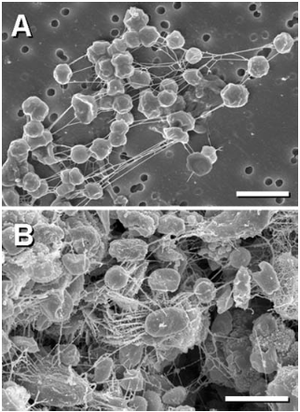
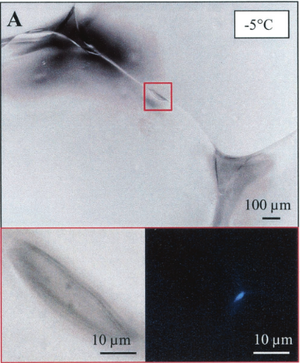
In environments where ice forms, new obstacles such as ice damage, oxidative insult, and osmotic imbalance (as reviewed in Siddiqui et al. 2013). PES to create biofilms that protect against ice crystal damage and form channels that allows nutrient transportation (as reviewed in Siddiqui et al. 2013). In a study examining samples of arctic sea ice, higher proportions of bacteria attached to particles (such as sediment grains, detritus, and ice crystal borders) were found as samples increased in temperature. The results from the study, aside from showing evidence for microbial communities which can live within a frozen environment, suggests bacterial attachment as a possible cold-adaptation mechanism (Junge et al. 2003). EPS, which favors cell attachment, might be the mechanism causing the increase rates of particle associated bacteria among the colder temperatures, as it is a known cold-adaptation mechanism (Junge et al. 2003).
At cold temperatures, H. lacusprofundi and M. burtonii both have changes in morphology and form multicellular aggregates, or packets of cells, which might be embedded in a EPS. These cell aggregates provide a survival advantage for cells because it facilitates exchange of nutrients, proteins, and genetic materials (Reid et al. 2006). Furthermore, both species were found in temperatures within the range of those found in Mars subsurface environments.
Colder temperatures also decrease membrane fluidity. One simple way of counteracting this effect is by increasing the proportion of unsaturated to saturated lipids in the membrane (as reviewed in Siddiqui et al. 2013). Exiguobacterium sibiricum has higher fatty acid desaturase gene expression at low temperatures, removes two hydrogen bonds from fatty acid (as reviewed in Siddiqui et al. 2013). M burtonii lacks the desaturase, instead alters expression of lipid biosynthesis genes, making fewer saturated lipids. Metagenomics show that microbes in glacier ice tended to have more genes involved in membrane maintenance (as reviewed in Siddiqui et al. 2013).
Halophiles and Methanogens
Halophiles and methanogens are among the top candidates as model organisms due to the fact that high salt water lowers the freezing temperature of water (DasSarma 2006). Such is the case in Deep Lake in Antarctica, which does not freeze despite having surface temperatures of -20 C (Dassarma 2006).
Methanogens are also good model organisms since they can grow in anoxic environments and signs of methane in the Mars atmosphere gathered from recent expeditions might have been produced by methanogens (Reid et al. 2006). Methanogens that thrive mainly on hydrogen were found in a 2002 experiment in Lidy Hot Springs, Idaho (Chapelle et al. 2002). This archaea-dominated community, which lives 200m under the surface, is important because it is a model for life on Mars or Europa mainly because of the lack of organic carbon in its environment. Using the hydrogen provided by the reaction of hydrothermal fluids reacting with surrounding rocks and carbon dioxide, the methanogens conduct the reaction 4H2 + CO2 -> CH4 + 2H2O for metabolism (Chapelle et al. 2002).
One of the most studied halophiles is Halobacterium sp. NRC-1, which can withstand a variety of environmental extremes including radiation, temperature, and heavy metal exposure. Halobacterium sp. NRC-1 has a variety of mechanisms to counteract solar radiation, including the production of cyclopyrimidine photolyase (phr) which can break down products caused by UV radiation. Cells under radiation stress may also begin producing membrane pigments to protect the cellular DNA or induce the gene rfa3 which encodes for a protein which may function in DNA repair (DasSarma 2006). Aside from its survival mechanisms, halophiles are also good models for interplanetary transport as they can live within protective salt crystals in meteorites. Salt crystals have been found in Martian meteorites Shergotty and Nahkla, indicating a precedence for a possible halophile environment in interspace travel (DasSarma 2006).
Radiation Damage
Radiation poses the most danger because of its damage to nucleic acids and consequently DNA. One of the most durable organisms in the presence of radiation is the bacterium Deinococcus radiodurans. This bacterium has been isolated from various dry and low-nutrient environments, suggesting an adaptation to dessication (as reviewed by Battista 1997). Indeed, resistance to dessication, which also can damage DNA, is usually correlated with increased radiation resistance. In one experiment, strains of D. radiodurans which were ionizing radiation-sensitive also showed sensitivity to dessication when left in a desiccator for six weeks (as reviewed by Battista 1997). Considering that there are not many natural environments on earth that produce extreme radiation, it is likely that D. radiodurans' radiation resistance is a byproduct of its adaptation to dessicaition. Similar connections between dessication and radiation resistance have been made in regard to tardigrades' ability to survive in space.
D. radiodurans can survive upwards of 17,000 gamma rays, compared to the lethal amounts of 10 gamma rays for humans (Talbott 2013). D. radiodurans, rather than passively resist radiation damage, uses various DNA repair mechanisms to repair the extensive DNA damage following radiation exposure (as reviewed by Battista 1997). Nonetheless, an important aspect of D. radiodurans' ability to repair itself is that it is multigenomic, containing four copies of its chromosome. Having multiple copies of chromosomes has been shown as being as improving radiation resistance in other organisms such as Saccharomyces cerevisiae (as reviewed by Battista 1997). Radiation-induced double-strand breaks (dsbs) in the DNA, among the more harmful of radiation effects, occurs randomly along the chromosome. By having multiple copies of the chromosome, there is a lower chance that damage occurs in the same place across each of the bacterium's copies. Through recombination of regular sections of DNA with damaged sections D. radiodurans is able to repair its genome once conditions are favorable to growth (as reviewed by Battista 1997).
Conclusion
References
DasSarma S. 2006. Extreme Halophiles Are Models for Astrobiology. Microbe / Volume 1, Number 3, 2006
Rothschild LJ, Mancinelli RL. 2001. Life in extreme environments. Nature 409(6823):1092-101
Talbott SL. 2013. Shattering the genome. In Context #30
Edited by student of Joan Slonczewski for BIOL 238 Microbiology, 2009, Kenyon College.