Human Astrovirus: Difference between revisions
| Line 28: | Line 28: | ||
==Description and Significance== | ==Description and Significance== | ||
[[File:humanastro.jpg|right|125px | [[File:humanastro.jpg|right|frame|125px|astrovirus]] | ||
Human Astrovirus (HAstV) is a species of the genera ''Mamastrovirus'', which are a group of enteric viruses known to be capable of causing mild symptomatic gastrointestinal infection. HAstV was first discovered as a causative agent for gastrointestinal infection among children under the age of 2. HAstV was first discovered in 1975, but the ''Astroviridae'' family was not established until 1995 [1]. In the early 2000s, three distinctive clades were discovered among the astrovirus species; VA1-VA3, VA2-VA4, and MLB [2]. | Human Astrovirus (HAstV) is a species of the genera ''Mamastrovirus'', which are a group of enteric viruses known to be capable of causing mild symptomatic gastrointestinal infection. HAstV was first discovered as a causative agent for gastrointestinal infection among children under the age of 2. HAstV was first discovered in 1975, but the ''Astroviridae'' family was not established until 1995 [1]. In the early 2000s, three distinctive clades were discovered among the astrovirus species; VA1-VA3, VA2-VA4, and MLB [2]. | ||
Revision as of 18:02, 22 April 2024
Classification
Higher Order Taxonomy
Virus
Realm: Riboviria
Kingdom: Orthornavirae
Phylum: Pisuviricota
Class: Stelpaviricetes
Order: Stellavirales
Family: Astroviridae
Genus: Mamastrovirus
Species
Human Astrovirus 1-8
Description and Significance
Human Astrovirus (HAstV) is a species of the genera Mamastrovirus, which are a group of enteric viruses known to be capable of causing mild symptomatic gastrointestinal infection. HAstV was first discovered as a causative agent for gastrointestinal infection among children under the age of 2. HAstV was first discovered in 1975, but the Astroviridae family was not established until 1995 [1]. In the early 2000s, three distinctive clades were discovered among the astrovirus species; VA1-VA3, VA2-VA4, and MLB [2].
Among other viruses, HAstV is notable for its resilience outside of a host environment. It has been determined that HAstV is resistant to inactivation by alcohols (including the common antiseptic ethanol), bleach, detergents, and to mild UV or heat treatments, along with being capable of staying alive in water for up to 90 days [3]. This makes HAstV commonplace in human environments, and as such HAstV has been found to responsible for roughly 4.0-8.6% of all diarrhea-related symptoms in young children [2]. Symptoms are relatively mild compared to other pathogens such as norovirus and rotavirus. Milder symptoms and the lack of prevalence in healthy young adults suggests that adaptive immunity is often developed in youth following a first encounter with HAstV [3].
Additionally of note are a few isolated cases associated with the HAstV serotype VA1. In 10 distinct cases, infection of the central nervous system has been noted, along with a lack of gastroenteric infection. Over half of the resulting cases were fatal [4]. It is unknown by what mechanisms these infections occur, although mink and ovine astrovirus infections have also resulted in CNS disease.
Genome Structure
Human Astrovirus has a single stranded, RNA-based genome, as is typical of many viruses. The genome is approximately 6.8kb long, and contains three main open reading frames (ORFs), known as ORF1a, ORF1b, and ORF2. ORF1a and ORF1b overlap one another by roughly 70 nucleotides, but the function of the five encoded peptides is relatively unknown. ORF2 is more well understood. ORF2 is roughly 2.4 kb long, and contains subgenomic RNA (sgRNA). The sgRNA allows for the encoding of multiple proteins over a single strand of RNA, and allows the genome to be more easily compressed. It is believed that ORF2 encodes for the capsid protein, and sgRNA allows for heavily increased expression of the capsid protein later in the replication cycle to accommodate for creation of new viruses [2].
The G+C content of the genome has not been analyzed, nor has it been sequenced and compared to other viruses. Since HAstV generates nucleotide mutations at a rate of 3.7 x 10^-3 substitutions per site per year [1], and astroviruses are so prevalent on a global scale, it is difficult to develop a definitive genome when so many different serotypes exist.
Cell Structure, Metabolism and Life Cycle
Cell Structure
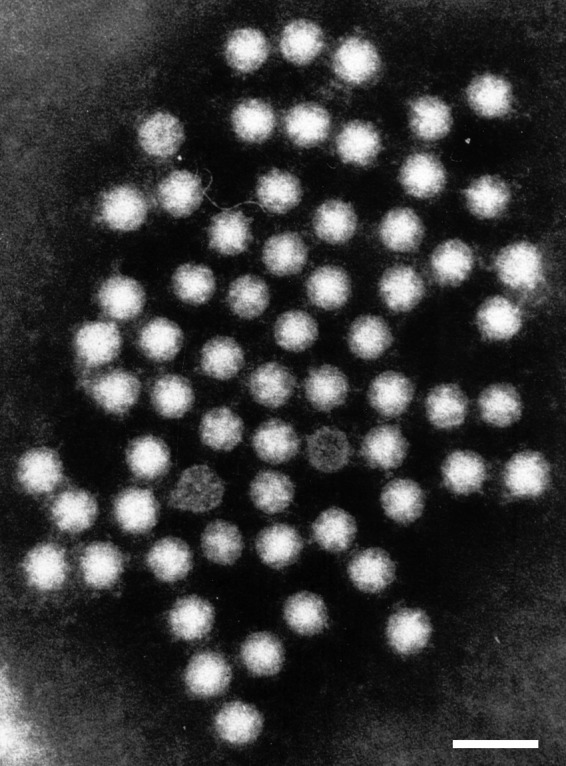
Human Astrovirus has a fairly simple structure, and one that is typical of other pathogenic viruses, such as SARS-CoV-2. It is a smooth, spherical, capsid-based virus that may have star-like projections [5]. The star-like projections are formed from a dimer of proteins contained within their genome. Only about 10% of replicated astroviruses have these star projections [1]; the other 90% have a more smooth, rounded surface. Astroviruses with spikes have a distinct pathogenic advantage in that they can use these spikes to attach to polysaccharides; upon attaching to the polysaccharide, receptor-mediated endocytosis allows the virus free passage into the cell [1]. The capsid is typically cleaved once outside the host cell to signify virus maturation.
Metabolism
Viruses have no cellular machinery and do not replicate on their own to survive. As such, they have no metabolism and do not uptake resources from their extraneous environment. Astroviruses instead make use of host cellular machinery, typically the rough endoplasmic reticulum, in order to replicate and continue to spread.
Life Cycle
Ecology and Pathogenesis
Habitat; symbiosis; biogeochemical significance; contributions to environment.
If relevant, how does this organism cause disease? Human, animal, plant hosts? Virulence factors, as well as patient symptoms.
References
Author
Page authored by Ethan Haggin and Kasia Jackowski, students of Prof. Jay Lennon at Indiana University.