Parvovirus B19: Difference between revisions
| Line 12: | Line 12: | ||
[[Image:Screenshot_2024-04-06_at_4.00.04_PM.png|thumb|300px|right| The V9 (A) and A6 (B) genomes are depicted with transcriptional landmarks. These include promoters (ie. P6), splice donors (D), and splice acceptors (A), as well as the ORFs encoded and their predicted protein sizes in kDa. Photo credit: [https://www-sciencedirect-com.libproxy.kenyon.edu/science/article/pii/S0042682209005303?via%3Dihub]]] | [[Image:Screenshot_2024-04-06_at_4.00.04_PM.png|thumb|300px|right| The V9 (A) and A6 (B) genomes are depicted with transcriptional landmarks. These include promoters (ie. P6), splice donors (D), and splice acceptors (A), as well as the ORFs encoded and their predicted protein sizes in kDa. Photo credit: [https://www-sciencedirect-com.libproxy.kenyon.edu/science/article/pii/S0042682209005303?via%3Dihub]]] | ||
The <i>Parvovirus</i> genome is a single strand of DNA with 5,596 nucleotides, 4,830 of which are coding regions.<ref name=ncbi/> This region contains 2 large open reading frames.<ref name=ncbi/> One large non-structural protein is coded by | The <i>Parvovirus</i> genome is a single strand of DNA with 5,596 nucleotides, 4,830 of which are coding regions.<ref name=ncbi/> This region contains two large open reading frames.<ref name=ncbi/> The genome of typical parvoviruses contain two promoters; one on the left for the non-capsid protein, and one in the middle for the two capsid proteins.<ref name=Ozawa>[https://journals-asm-org.libproxy.kenyon.edu/doi/10.1128/jvi.61.8.2395-2406.1987. Ozawa, K., Ayub, J., Hao, Y.S., Kurtzman, G., Shimada, T., and Young, N. "Novel transcription map for the B19 (human) pathogenic parvovirus" 1987. Journal of Virology, 61(8), 2395–2406.]</ref> The B19 parvovirus produces all transcripts starting from a singular promoter on the left side of the genome.<ref name=Ozawa/> There were four RNA species that appeared to have splicing behaviors, and three RNA species terminate in the middle of the genome.<ref name=Ozawa/> The image to the right shows the genome for prototype B19V isolates, and includes the four spliced RNA species, c, d, g, and h.<ref name=Ozawa/> Species c consists of two exons of 0.3 and 1.95 kb, d consists of two exons of 0.2 and 1.95 kb, g consists of two 0.3 kb exons, and h consists of two exons of 0.2 and 0.3 kb.<ref name=Ozawa/> | ||
The genome has two large open reading frames.<ref name=ncbi/> One large non-structural protein is coded by the left open reading frame, NS1, and the second reading frame codes for two capsid proteins, VP1 and VP2.<ref name=ncbi/><ref name=Ozawa/> The sequences of B19V isolates do not exhibit much genetic variation with NS1 showing incredibly high conservation, and 2-3% divergence in VP1 and VP2 regions.<ref name=ncbi/> When examining isolates from patients with chronic infection due to B19V, there is a much higher degree of variation at both the DNA and protein level.<ref name=Hemauer/> | |||
[[Image:Screenshot_2024-04-06_at_4.00.04_PM.png|thumb|300px|right| The V9 (A) and A6 (B) genomes are depicted with transcriptional landmarks. These include promoters (ie. P6), splice donors (D), and splice acceptors (A), as well as the ORFs encoded and their predicted protein sizes in kDa. Photo credit: [https://www-sciencedirect-com.libproxy.kenyon.edu/science/article/pii/S0042682209005303?via%3Dihub]]] | |||
There are now 3 distinct genotypes recognized as <i>Parvovirus B19</i> including: (1) all prototype B19V isolates, (2) A6 and LaLi isolates, and (3) V9 and related isolates.<ref name=Chen/> These variations in the genome are potentially responsible for the variety of host responses to infection.<ref name=Chen/> In a study on 3 isolates of the 3 genotypes, the A6 isolate did not produce R5, R7, and R9 mRNAs, which are important in the production of VP1 and VP2 as coordinating mRNAs.<ref name=Chen/> There was also an increase in sequence divergence between the 3 isolates. They found that B19V NS1-V9 and B19V NS1-A6 diverge by 13% from B19V NS1, but only diverge by 6% in protein structure.<ref name=Chen/> | There are now 3 distinct genotypes recognized as <i>Parvovirus B19</i> including: (1) all prototype B19V isolates, (2) A6 and LaLi isolates, and (3) V9 and related isolates.<ref name=Chen/> These variations in the genome are potentially responsible for the variety of host responses to infection.<ref name=Chen/> In a study on 3 isolates of the 3 genotypes, the A6 isolate did not produce R5, R7, and R9 mRNAs, which are important in the production of VP1 and VP2 as coordinating mRNAs.<ref name=Chen/> There was also an increase in sequence divergence between the 3 isolates. They found that B19V NS1-V9 and B19V NS1-A6 diverge by 13% from B19V NS1, but only diverge by 6% in protein structure.<ref name=Chen/> | ||
==B19V's Essential Proteins== | ==B19V's Essential Proteins== | ||
Revision as of 22:10, 12 April 2024
Background
By Grace Potter
Parvovirus B19 is the only member of the Parvoviridae family that has been found to infect human hosts.[1] It was discovered in 1974, when a research group looking at hepatitis B surface antigens found a serum sample with unexpected results.[1] Another lab in Japan described a similar virus in 1979 that they called "Nakatami".[1] When compared, the two were found to be identical.[1] In 1985 this virus was officially recognized as a member of the Parvoviridae family due to its similarities in genome size and density.[1]
Infection by Parvovirus B19 (Parvo B19V) causes many diseases, including "fifth disease" in children, aplastic crisis for people with hemolytic anemia, anemia in immunocompromised patients, acute or chronic arthropathy in adults, and fetal hydrops in pregnant women.[2][3] Changes in the genome are potentially responsible for the wide variation of clinical presentations associated with B19V infection.[2]
Parvoviruses are classified in reference to what type of host they are capable of infecting and how they reproduce.[1] Those capable of infecting vertebrate hosts are referred to as "Parvovirinae" and are further subdivided by reproduction processes.[1] "Parvovirus" refers to members of the Parvoviridae family that reproduce autonomously, but there are also members of this family seen reproducing with a helper virus, "Dependovirus", and preferentially in erythroid cells, "Erythrovirus".[1] Parvovirus B19 has recently been found to exhibit extreme specificity for human erythroid progenitor cells, and has therefore been classified as an "erythrovirus".[4][1]
Genome Structure

The Parvovirus genome is a single strand of DNA with 5,596 nucleotides, 4,830 of which are coding regions.[1] This region contains two large open reading frames.[1] The genome of typical parvoviruses contain two promoters; one on the left for the non-capsid protein, and one in the middle for the two capsid proteins.[5] The B19 parvovirus produces all transcripts starting from a singular promoter on the left side of the genome.[5] There were four RNA species that appeared to have splicing behaviors, and three RNA species terminate in the middle of the genome.[5] The image to the right shows the genome for prototype B19V isolates, and includes the four spliced RNA species, c, d, g, and h.[5] Species c consists of two exons of 0.3 and 1.95 kb, d consists of two exons of 0.2 and 1.95 kb, g consists of two 0.3 kb exons, and h consists of two exons of 0.2 and 0.3 kb.[5]
The genome has two large open reading frames.[1] One large non-structural protein is coded by the left open reading frame, NS1, and the second reading frame codes for two capsid proteins, VP1 and VP2.[1][5] The sequences of B19V isolates do not exhibit much genetic variation with NS1 showing incredibly high conservation, and 2-3% divergence in VP1 and VP2 regions.[1] When examining isolates from patients with chronic infection due to B19V, there is a much higher degree of variation at both the DNA and protein level.[3]

There are now 3 distinct genotypes recognized as Parvovirus B19 including: (1) all prototype B19V isolates, (2) A6 and LaLi isolates, and (3) V9 and related isolates.[2] These variations in the genome are potentially responsible for the variety of host responses to infection.[2] In a study on 3 isolates of the 3 genotypes, the A6 isolate did not produce R5, R7, and R9 mRNAs, which are important in the production of VP1 and VP2 as coordinating mRNAs.[2] There was also an increase in sequence divergence between the 3 isolates. They found that B19V NS1-V9 and B19V NS1-A6 diverge by 13% from B19V NS1, but only diverge by 6% in protein structure.[2]
B19V's Essential Proteins
Parvoviruses are among the smallest DNA containing viruses that are capable of infecting mammal hosts.[1] The virion consists of 3 proteins (NS1, VP1, and VP2) and both the positive and negative DNA strands of its singular chromosome.[1] The capsid has icosahedral symmetry, and includes 60 copies of the capsomer proteins VP1 and VP2.[1] VP2 makes up 96% of the capsid and is encoded from 3125 nt to 4786 nt. VP1 is the minor capsid protein encoded from 2444 nt to 4786 nt. The VP1 coding region is identical to VP2 except for an additional 227 amino acids referred to as the VP1 unique region, or VP1u.[1][6]
The major nonstructural protein coded for by B19V is NS1, which is encoded by nucleotides between 435 and 2448.[1] NS1 is thought to contain a DNA binding domain at the N-terminus with conserved motifs for single-strand nickase and an ATP/helicase domain in the center, and a transcription activation at the C-terminal.[2] For a long time the role of NS1 in infection was largely unknown, but recent studies have highlighted its potential in the proliferation of chronic symptoms related to Parvovirus B19V infection.[1][7]
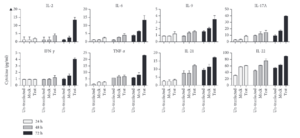
Results from a study indicate that the transfection of hPVB19 NS1 into HEK-293T cells increased proinflammatory cytokine levels.[7] This is consistent with the findings that NS1 might play a role in facilitating the upregulation of inflammatory reactions, including chronic arthritis which can be a symptom of chronic B19V infection.[7]
The figure to the right indicates a significant increase in the expression levels of the cytokines IL-17A and TNF-α. IL-17A and TNF-α initiate inflammation cascades in host cells.[7] Inflammation has been found to increase the risk of cells becoming carcinogenic, and further research is needed to assess the role of these cytokines in cancer development.[7] An increase in IFN and TNF in supernatants from NS1-transfected cells can induce the expression of IL-6, an important proinflammatory cytokine, and shows the potential of NS1 in triggering autoimmune disorders.[7] Further research is needed to fully understand the impact of NS1 on autoimmune disorders, especially considering its impact on proinflammatory cytokines.
Differences in the NS1 proteins found across the 3 genotypes of Parvovirus B19 show that B19V NS1-V9 and B19V NS1-A6 diverge by 13% from B19V NS1 in genetic sequence, but only diverge by 6% in protein structure.[2] The inability of B19V NS1-A6 to replicate the prototype B19V genome in comparison with its ability to replicate and package other A6 genomes, shows that the NS1 protein has genotype specific activity.[2]
Of the proteins produced by Parvovirus B19, only NS1 is predicted to possess a classical nuclear localization signal (cNLSL).[8] NS1 becomes detectable at low levels 6 hours post infection, and peaks 24 to 36 hours post infection.[8] Confocal laser scanning microscopy confirmed that NS1 accumulated in the nucleus.[8] More on the nuclear localization of NS1 in the pathogenesis and infectivity section.
In many parvoviruses, the NS1 protein contains a cNLS sequence.[8] Some of the cNLSs are more similar to each other than others, and overall these DNA and amino acid sequences are not highly conserved.[8] The parvovirus minute virus of mice and ungulate protoparvovius 1 both have bipartite cNLSs, but human parvovirus B19 has a monopartite cNLS.[8] Bipartite cNLSs are more efficient at binding the IMP nuclear translocation protein, so the presence of a monopartite cNLS could be an effort to limit NS1 levels in the nucleus.[8] This seems to provide evidence for the independent evolution of cNLS sequences, and therefore the importance of the NS1 protein entering the nucleus in the viral life cycle.[8]
There are other open reading frames in the B19V genome, but the function of their resulting proteins is largely unknown.[1] Further research is needed in this area.
Pathogenesis and Infection
B19V has an extremely high selectivity and cell-type tropism for human erythroid progenitor cells.[4] Infection of endothelial cells is extremely unlikely in genotype 1 B19V's, but has been seen in genotype 2.[4][2] It has been found that B19V can only successfully replicate in bone-marrow erythroid progenitor cells, or conditions that mirror this setting.[2][9]
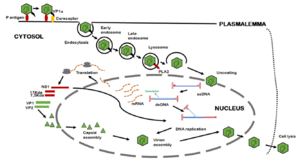
B19V's path of infection starts with entrance through the respiratory tract, and from there it reaches the bloodstream and eventually the bone marrow.[9] Interactions between the bone-marrow erythroid progenitor cells glycolipid globosides, blood group P antigen, and an unknown secondary receptor allow the viral particle to enter the cell.[9] The interaction between the viral shell and P antigen leads to the externalization of the unique region of VP1, which consequently allows B19V to enter the cell through an endocytic pathway.[9] The VP1u region exhibits phospholipase A2 activity, which allows the virus to evade lysosomal fusion and ensure infectivity when it is exposed.[9]
As the infection continues, apoptosis and depletion of infected cells leads to erythropoiesis arrest, erythroid aplasia, and the appearance of giant erythroblasts.[9] The replication and release of B19V can lead to viremia, with increased viral loads of 1012 virus/mL.[9] Infection is normally asymptomatic, but patients with chronic illness, immunodeficiency, and pregnancy can experience much more severe symptoms.[9]
B19V is recognized by pathogen-associated molecular pattern molecules and pattern recognition receptors, and exhibits typical prodromal phase production of cytokines.[9] There is no imbalance of cytokine patterns in persistent infection, except the cytokine IFN shows increased expression in long-term infection.[9] B19V inhibits erythroid cell growth and creates a pile-up of cells in S, G2, and M phases that then go through apoptosis.[9]
Antibodies specific to B19V are produced early in infection and typically neutralize symptoms.[9] IgM is produced 8-12 days post infection and remains present for 3-6 months, and IgG antibodies are produced a couple of days after IgM production begins.[9] As IgM concentrations decrease, IgG antibodies stay present in the body.[9] The majority of B19V antibodies are directed against the structural proteins VP1 and VP2.[9] Interestingly, antibodies against NS1 protein have been found in 30% of subjects with recent infections, and in those with chronic infections.[9]
One very important aspect of B19V pathogenesis is the nuclear import of NS1. Because NS1 is too large to enter through passive diffusion, the IMPα/β nuclear transport protein is necessary.[8] Alvisi et al (2023) designed an experiment to test this using NS1 wild type proteins, an NS1 protein with a K177T substitution, and a mini-genome without the start codon for NS1 in EPO-S1 cells.[8] NS1 proteins were detected 24 hours post transfection in the wild type and K177T substitution cells, but not in cells without the start codon.[8] NS1 was found in the nucleus of cells that had been transfected with wild type NS1, but the K177T substitution NS1 ended up in the cytosol.[8] Viral transcription was found in the wild type cells 24 and 48 hours post transfection, but not in the K177T substitution cells.[8] More research is needed to fully understand the importance of NS1's nuclear location, but it appears to be heavily linked to the ability of the virus to replicate.
NS1 has also been found in the cytosol of many B19V infected cells. A common substitution, K177C, is unable to translocate to the nucleus, but appears to retain the ability to induce apoptosis.[8] Therefore, there is a possible functional role of NS1 in the cytoplasm.
Potential Use in Development of New Treatments
Conclusion
References
- ↑ 1.00 1.01 1.02 1.03 1.04 1.05 1.06 1.07 1.08 1.09 1.10 1.11 1.12 1.13 1.14 1.15 1.16 1.17 1.18 1.19 1.20 Heegaard, E.D. and Brown, K.E. "Human Parvovirus B19." 2002. Clinical Microbiology Review 15(3):485-505.
- ↑ 2.00 2.01 2.02 2.03 2.04 2.05 2.06 2.07 2.08 2.09 2.10 Chen, Z., Guan, W., Cheng, F., Chen, A.Y., and Qiu, J. "Molecular characterization of human parvovirus B19 genotypes 2 and 3" 2009. Virology, 394(2), 276-285.
- ↑ 3.0 3.1 Hemauer, A., von Poblotzki, A., Gigler, A., Cassinotti, P., Siegl, G., Wolf, H., and Modrow, S. "Sequence variability among different parvovirus B19 isolates" 1996. Journal of General Virology, 77(8), 1781-1785.
- ↑ 4.0 4.1 4.2 Rinkūnaitė, I., Šimoliūnas, E., Bironaitė, D., Rutkienė, R., Bukelskienė, V., Meškys, R., and Bogomolovas, J. "The Effect of a Unique Region of Parvovirus B19 Capsid Protein VP1 on Endothelial Cells" 2021. Biomolecules, 11(4), 606.
- ↑ 5.0 5.1 5.2 5.3 5.4 5.5 Ozawa, K., Ayub, J., Hao, Y.S., Kurtzman, G., Shimada, T., and Young, N. "Novel transcription map for the B19 (human) pathogenic parvovirus" 1987. Journal of Virology, 61(8), 2395–2406.
- ↑ Ros, Carlos, Bieri, Jan, and Leisi, Remo. "The VP1u of Human Parvovirus B19: A Multifunctional Capsid Protein with Biotechnological Applications" 2020. Viruses, 12(12), 1463.
- ↑ 7.0 7.1 7.2 7.3 7.4 7.5 7.6 Jalali, Sedigheh, Farhardi, Ali, Dehbidi, G.R., Farjadian, Shirin, Sharifzadeh, Sedigheh, Ranjbaran, Reza, Seyyedi, Noorossadat, Namdari, Sepide and Behzad-Behbahani, Abbas. "The Pathogenic Aspects of Human Parvovirus B19 NS1 Protein in Chronic and Inflammatory Diseases" 2022. Interdisciplinary Perspectives on Infectious Diseases.
- ↑ 8.00 8.01 8.02 8.03 8.04 8.05 8.06 8.07 8.08 8.09 8.10 8.11 8.12 8.13 Alvisi, G., Manaresi, E., Cross, E.M., Hoad, M., Akbari, N., Pavan, S., Ariawan, D., Bua, G., Petersen, G.F., Forwood, J., and Gallinella, G. "Importin α/β-dependent nuclear transport of human parvovirus B19 nonstructural protein 1 is essential for viral replication" 2023. Antiviral Research, 213.
- ↑ 9.00 9.01 9.02 9.03 9.04 9.05 9.06 9.07 9.08 9.09 9.10 9.11 9.12 9.13 9.14 9.15 9.16 Zakrzewska, K., Arvia, R., Bua, G., Margheri, F., and Gallinella, G. "Parvovirus B19: Insights and implication for pathogenesis, prevention and therapy" 2023. Aspects of Molecular Medicine.
Authored for BIOL 238 Microbiology, taught by Joan Slonczewski,at Kenyon College,2024