Styrene metabolism in bacteria
Styrene and polystyrene
Styrene is an aromatic compound with many industrial uses. It is the building block of polystyrene, which is used in various consumer and industrial goods ranging from foam containers and packaging material (commonly known as "Styrofoam") to plastic containers such as CD cases and Petri dishes.
Although styrene and its derivatives are useful, they pose significant environmental problems as styrene has been found to be toxic 11. Current industrial methods for producing styrene are energetically expensive[2,3], and industrial uses of styrene often release styrene into the environment. Polystyrene is highly recalcitrant and persists in the environment, and is usually disposed of by incineration or accumulation in landfills. Styrene and polystyrene pose problems in energy consumption, pollution and waste management, so there is great interest in using microorganisms as inexpensive tools for addressing these problems.
Styrene catabolism
There many bacteria that can use styrene as an energy and carbon source, but most research has focused on a few species, namely Pseudomonas putida, Rhodococcus opacus and Pseudomonas fluorescens. Styrene catabolism is dependent on oxygen availability, and occurs in the absence of more favourable catabolic substrates.
Mechanism of degradation
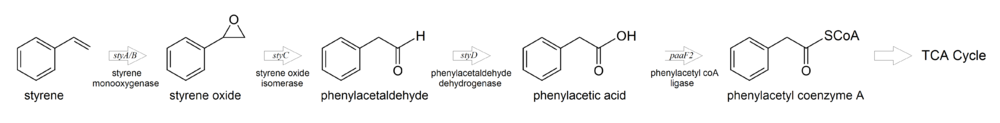
The most commonly described pathway for styrene degradation involves the oxidation of styrene to phenylacetate, which is then converted to TCA cycle intermediates. This pathway is shown below:
The steps leading from styrene to phenylacetate are collectively known as the "upper pathway" of styrene degradation, and the steps starting from phenylacetate are known as the "lower pathway" of styrene degradation[1].
In the upper pathway, styrene is first oxidized to styrene oxide by styrene monooxygenase, which requires a reductant for electrons. Styrene oxide is then rearranged to phenylacetaldehyde by styrene oxide isomerase, and further oxidized by phenylacetaldehyde dehydrogenase to produce phenylacetate. In the lower pathway, phenylacetate is ligated to coenzyme A to produce phenylacetyl coA, and then split by several enzymes to produce acetyl coA, which enters the TCA cycle.
Catabolic enzymes
The upper pathway enzymes are encoded by the styABCD operon in strains of Pseudomonas and Rhodococcus.
The styA and styB genes encode the styrene monooxygenase complex. StyA has styrene monooxygenase activity and converts styrene to styrene oxide. It requires electrons from FADH2. StyB has FAD reductase activity and transfers electrons from NADH to FAD+ to supply FADH2 for StyA.
In some strains of Rhodococcus, styA and styB have been combined into a single gene, styA2B, which encodes an enzyme that possesses both styrene monooxygenase and FAD reductase activities and is more efficient than the StyA/StyB styrene monooxygenase complex.
styC encodes the styrene isomerase enzyme, and styD encodes the phenlyacetaldehyde dehydrogenase enzyme.
Finally, another gene, styE, has been observed in some strains of Pseudomonas[6]. It appears to encode an ATP-dependent styrene transporter, and is part of the same operon as the styABCD genes.
The lower pathway of styrene degradation is initiated by phenylacetate coenzyme A ligase. Pseudomonas putida possesses three genes for this enzyme, of which one gene, paaF2, is transcribed in the presence of styrene.
Regulation of styrene degradation
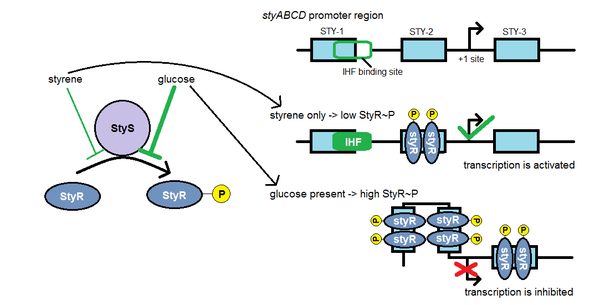
Styrene catabolism is promoted in the presence of styrene, and repressed in the presence of more favourable nutrients like glucose. The styABCD(E) operon is regulated by a two-component system, the StyS/StyR system.
The StyS protein is a sensor kinase that phosphorylates the response regulator, StyR, in response to styrene concentration. StyS has been shown to have two sensor domains and two kinase domains, which suggests that it may respond to two different signals. In addition to styrene, the other stimulus is hypothesized to be a high redox potential (indicating the presence of a more energetically favourable substrate such as glucose).
In the presence of styrene, StyS phosphorylates StyR at a low level, resulting in low levels of StyR~P, which promotes transcription of the operon. However, in the presence of a more favourable metabolite such as glucose, StyS phosphorylates StyR more rapidly, resulting in high levels of StyR~P, which inhibits transcription of the operon. In this way, styrene degrading bacteria can control their use of styrene so that they only degrade it in the absence of more favourable metabolic substrates.
Biotechnical applications of styrene catabolism
Styrene-degrading bacteria could potentially be used in bioremediation of styrene pollution. A biofilter consisting of Brevibacillus sp. has been shown to remove 3 kg of styrene in a day[7], and Rhodococcus ruber has been shown to form biofilms on polystyrene and partially degrade it[8].
A team of researchers has also devised a two-step process for the conversion of polystyrene into polyhydroxylalkanoates, by converting polystyrene into styrene oil using pyrolysis and adding it to a fermentor containing Pseudomonas putida[9][10]. Polyhydroxylaalkanoates are naturally synthesized by bacteria in response to stress, and can be used as a biodegradable thermoplastic.[1]
The ability to use bacteria to degrade styrene and convert recalcitrant polystyrene into an environmentally friendly plastic has great potential for addressing the environmental problems posed by styrene and polystyrene.
References
- Mooney A., Ward P.G. and O’Connor K.E. “Microbial degradation of styrene: biochemistry, molecular genetics, and perspective for biotechnical applications.” Applied Microbiology and Biotechnology, 2006.
- Adkins J., Pugh S., McKenna R., and Nielsen D.R. “Engineering microbial chemical factories to produce renewable ‘biomonomers’.” Frontiers in Microbiology, 2012. 3, Article 313.
- US Department of Energy. “Steam System Opportunity Assessment for the Pulp and Paper, Chemical Manufacturing, and Petroleum Refining Industries.” US Department of Energy, 2002. Retrieved from http://www1.eere.energy.gov/manufacturing/tech_deployment/pdfs/steam_assess_mainreport.pdf on 15 November 2012.
- Mooney A., O’Leary N.D., and Dobson A.D. “Cloning and functional characterization of the styE gene, involved in styrene transport in Pseudomonas putida CA-3.” Applied and environmental microbiology, 2006. 72(2), 1302 – 1309.
- Rampioni G., Leoni L., Pietrangeli B., and Zennaro E. “The interplay of StyR and IHF regulates substrate-dependent induction and carbon catabolite repression of styrene catabolism genes in Pseudomonas fluorescens ST.” BMC (BioMed Central) microbiology, 2008. 8, 92.
- Leoni L., Rampioni G., Di Stefano V., and Zennaro E. “Dual role of response regulator StyR in styrene catabolism regulation.” Applied environmental microbiology, 2005. 71(9), 5411 – 5419.
- Hwang J.W., Choi C.Y., Park S., Lee E.Y. “Biodegradation of gaseous styrene by Brevibacillus sp. using a novel agitating biotrickling filter.” Biotechnology letters. 30(7), 1207 – 1212.
- Mor R., Sivan A. “Biofilm degradation and partial degradation of polystyrene by the actinomycete Rhodococcus ruber: Biodegradation of polystyrene.” Biodegradation, 2008. 19(6), 851 – 858.
- Ward P.G., Goff M., Donner M., Kaminsky W. and O’Connor K.E. “A two step chemo-biotechnical conversion of polystyrene to a biodegradable thermoplastic.” Environmental Science and Technology, 2006.
- Biello D. “Bacteria Turn Styrofoam into Biodegradable Plastic.” Scientific American, 27 February 2006. http://www.scientificamerican.com/article.cfm?id=bacteria-turn-styrofoam-i
- Liebman K.C. “Metabolism and toxicity of styrene.” Environmental Health Perspectives, 1975.