Swarming Motility of Pseudomonas aeruginosa: Difference between revisions
No edit summary |
No edit summary |
||
| Line 87: | Line 87: | ||
(15) Murray, S. Thomas and Barbara I. Kazmierczak. “Pseudomonas aeruginosa Exhibits Sliding Motility in the Absence of Type IV Pili and Flagella.” Journal of Bacteriology 190.8(2008): 2700–2708.Print. | (15) Murray, S. Thomas and Barbara I. Kazmierczak. “Pseudomonas aeruginosa Exhibits Sliding Motility in the Absence of Type IV Pili and Flagella.” Journal of Bacteriology 190.8(2008): 2700–2708.Print. | ||
Revision as of 07:50, 22 November 2013
Pseudomonas aeruginosa Overview
Pseudomonas aeruginosa (P. aeruginosa) is a Gram-negative opportunistic pathogen which can cause serious diseases in immunocompromised patients (eg. cystic fibrosis patients or severe burn patients)[12]. The remarkable ability of living on a variety of organic food sources enables P. aeruginosa to colonize different environments including water, soil, plant and animal tissues [11]. For example, P. aeruginosa can utilize quaternary amine compounds such as carnitine and short-chain acylcarnitines that are plentiful in animal tissues as sole carbon and nitrogen sources [5]. P. aeruginosa is reported as the most common hospital pathogen partially due to its ability to form biofilms on medical devices and its resistance to a wide range of antibiotics [2].
Swarming Motility Overview
Recent studies show that the pathogenesis and virulence of P. aeruginosa are greatly contributed by its four different kinds of motilities, namely, twitching motility, sliding motility, swimming motility and swarming motility (TABLE. 1) [6]. Swarming motility is defined as a fast coordinated bacteria movement across a semisolid surface under nitrogen limiting conditions in response to certain amino acids [6]. A typical swarming colony gives rise to dendritic-like patterns as a result of bacteria migration away from their inoculation point [9] (FIG. 1). Swarming motility can provide bacteria a competitive advantage in searching for nutrient-rich environments and it can be affected by bacterial population size, water content and nutrient composition [7]. Since P. aeruginosa swarmer cells show significant differences in their morphology, pathogenic gene expression, antibiotic resistance and other adaptation strategies comparing to non-swarmer cells, swarming is classified as a flaxible lifestyle altered by environmental stimuli [12].
Swarming-associated Phenotype
Hyperflagellation
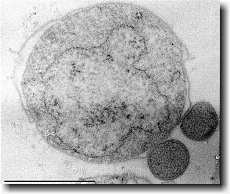
Compared to normal P. aeruginosa cells, which have single polar flagellum, swarming cells are hyperflagellated expressing two polar flagella [12]. This morphological change can facilitate bacteria movement over semisolid surface by providing more propulsion. The appearance of flagellated cells can be observed at the leading edge of a growing colony using phase contract, fluorescent or electron microscopy [1]. Alternatively, a motor specified for generating extra moving force can be synthesized in P. aeruginosa living in a viscous environment [1].
Cell Elongation
The size of wild-type P. aeruginosa cells are typically 0.5 to 0.8 µm by 1.5 to 3.0 µm [13]. Swarmer cells are elongated compared to the vegetative cells (cells that are capable of actively growing) [12]. Cells at the tip region of the dendrite are approximately two times the length of the center cells [8]. This morphological differentiation may be due to the gene suppression of cell division in tendril-tip swarmer cells. However, no regulatory mechanism of cell division during swarming has been proposed at the molecular level [1].
Change in Gene Expression
The genome size of P. aeruginosa strain PA01 is around 6264404 base pairs. The majority of the genes are involved in cell movement, nutrients uptake, antibiotic resistance and environmental stimuli responding systems [11].
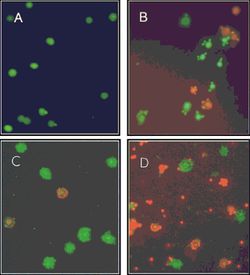
Further Microarrays study demonstrates that comparing the swarmer cells at the marginal region of a swarming colony to the bacteria grown under swimming condition, 7.5% of total P. aeruginosa genes (417 genes) from 21 distinguishable categories change more than two folds in their expression: 108 genes are down-regulated and 309 genes are up-regulated [7]. Genes related to virulence factors synthesis and substance export exhibit a great up-regulating tendency. For example, genes for the extracellular alkaline protease, type III secretion system (TTSS), and genes encoding redox-active phenazine compounds show 2 to 13 folds increase [7]. Interestingly, a different Microarray study focused on the distinct bacterial subpopulations within a P. aeruginosa PA14 strain swarming colony illustrates that cells at the fore end of the dendrite, compared to the center populations, promotes the expression of genes related to energy metabolism, ribosomal protein and down regulate the transcription of some of the virulence factors (eg. secreted factors and iron acquisition) (FIG.2) [9].
Potential Mechanism
The swarming motility in P. aeruginosa is known as a complicated adaptation process under semisolid surface conditions through altered gene expression and no conclusive mechanism of this physiological state has been proposed so far [7]. Several relevant studies demonstrate that the swarming motility is closely related to surfactant production [8] and quorum sensing [3].
Surfactant Production
The production of rhamnolipid is fundamental for the swarming motility as rhamnolipid is a surfactant facilitating the uptake and biodegradation of insoluble hydrocarbons which help to reduce surface tension and permit spreading motility [1][10]. In P. aeruginosa, the genes regulating the production of rhamnolipid are located at the rhl cell-to-cell signaling system [8]. When a few amino acids (eg. glutamate, aspartate, proline and histidine) that can promote rhamnolipid production serve as the sole nitrogen source, swarming motility is also up-regulated in wild-type strain, PT5 [8]. On the contrary, amino acids (eg. asparagine, glutamine and arginine) that suppress the synthesis of rhamnolipids can not induce swarming [8].
Quorum Sensing
When the amount of autoinducer (AHL) production in P. aeruginosa PAO1 is directly reduced by expressing the AHL-degrading gene (aiiA) homologue from Bacillus sp. A24, a significant down-regulation in several types of virulence factors and cytotoxic compounds together with a strongly reduced swarming are observed [3]. Alternatively, the swarming motility can be repressed indirectly by higher concentration of the culture agar (agar concentration > 0.5%) as harder surface limit the quorum sensing at the tip of the dendrite [4]. Further gene analysis emphasizes the necessity of quorum sensing for rhamnolipid production as the front edge of swarming tendril has high rhlA-gfp reporter expression under standard condition (agar concentration = 0.5%) while a lack of rhlA-gfp reporter expression close to the tip region is observed under harder surface condition (agar concentration > 0.5%) [4].
=References
(1) B. Kearns, B. Daniel. “A field guide to bacterial swarming motility.” Nature Reviews Microbiology 8(2010): 634-644. Print.
(2) Gellatly, L. Shaan and Robert E.W. Hancock. “Pseudomonas aeruginosa: new insights into pathogenesis and host defenses.” Pathogens and disease 67(2013): 159-173.Print.
(3) Reimmann, Cornelia et al. “Genetically programmed autoinducer destruction reduces virulence gene expression and swarming motility in Pseudomonas aeruginosa PAO1.”Microbiology 148 (2002): 923–932. Print.
(4) Kamatkar, G. Nachiket and Joshua D. Shrout. “Surface Hardness Impairment of Quorum Sensing and Swarming for Pseudomonas aeruginosa.”PLoS One 6.6(2011): 1-9. Print.
(5) Meadows, A. Jamie and Matthew J. Wargo. “Characterization of Pseudomonas aeruginosa Growth on O-Acylcarnitines and Identification of a Short-Chain Acylcarnitine Hydrolase.”Applied Environmental Microbiology 79.11(2013): 3355-3363. Print.
(6) Overhage, Joerg et al. “Identification of Genes Involved in Swarming Motility Using a Pseudomonas aeruginosa PAO1 Mini-Tn5-lux Mutant Library.” Journal of Bacteriology 189.5 (2007): 2164-2169. Print.
(7) Overhage, Joerg et al. “Swarming of Pseudomonas aeruginosa Is a Complex Adaptation Leading to Increased Production of Virulence Factors and Antibiotic Resistance.” Journal of Bacteriology 190.8 (2008):2671-2679. Print.
(8) THILO KO¨HLER et al. “Swarming of Pseudomonas aeruginosa Is Dependent on Cell-to-Cell Signaling and Requires Flagella and Pili.” Journal of Bacteriology 182.21 (2000): 5990–5996. Print.
(9) Tremblay, Julien and Eric Déziel. “Gene expression in Pseudomonas aeruginosa swarming motility.”BMC Genomics11.587 (2010):1-15. Print.
(10) Vatsa, Parulet al. “Rhamnolipid Biosurfactants as New Players in Animal and Plant Defense against Microbes.”International Journal of Molecular Science 11.12(2010): 5095–5108.Web. 31 Oct. 2013.
(11) www.pseudomonas.com
(12) Yeung, T. Y. Amy et al. “Swarming of Pseudomonas aeruginosa Is Controlled by a Broad Spectrum of Transcriptional Regulators, Including MetR.” Journal of Bacteriology 191.18(2009): 5592-5602. Print.
(13) Todar, Kenneth. “Pseudomonas aeruginosa.” Todar’s online textbook of bacteriology. N.p.n.d.Web.15 Nov.2013.
(14) Henrichsen, J¢rgen. “Twitching Motility.”Annual review of microbiology 37(1983):81-93.Print.
(15) Murray, S. Thomas and Barbara I. Kazmierczak. “Pseudomonas aeruginosa Exhibits Sliding Motility in the Absence of Type IV Pili and Flagella.” Journal of Bacteriology 190.8(2008): 2700–2708.Print.