Uranium Bioremediation in Contaminated Groundwater: Difference between revisions
No edit summary |
No edit summary |
||
| Line 1: | Line 1: | ||
{{Uncurated}} | |||
=General Introduction= | =General Introduction= | ||
[[File:wikipic1.png|300px|thumb|right|One of the many sites where soil and groundwater are contaminated by uranium. | [[File:wikipic1.png|300px|thumb|right|One of the many sites where soil and groundwater are contaminated by uranium. | ||
Latest revision as of 15:51, 2 October 2015
General Introduction

Uranium, an element known for its radioactivity and chemical toxicity is the forty ninth most abundant element that can be found in the Earth’s crust [1]. The abundance of this element has led to many years of usage such nuclear research, manufacturing of nuclear weapons and nuclear fuels. This led to extremely high amounts of uranium being released into the environment and has a high potential of damaging ecological systems. Additionally, the uranium can be secreted in an oxidized form [i.e., U(VI)] which is soluble and mobile resulting in contamination of groundwaters [2]. This led to the contamination of water supplies. To solve this problem, uranium bioremediation methods using certain types of microorganisms were created because unlike toxic organic compounds, degradation of toxic metals is not possible [1]. Currently, microorganisms are used to perform oxidation-reduction reactions on U(VI) to make it less soluble and therefore immobilizing it [3]. The immobilized uranium would then be removed from the groundwater.
Physical Environment
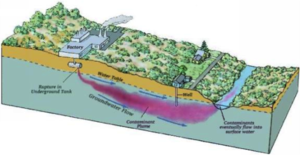
Uranium contaminated groundwater is usually found where there have been uses of uranium for research or production of weapons and fuels [1]. Since soluble U(VI) is mobile, it is able to contaminate aquifers which results in a widespread of groundwater contamination.
Oxygen
The process of uranium bioremediation requires that the soluble U(VI) is used as an electron acceptor and be reduced to insoluble U(IV). This is known as anaerobic respiration. The environment of groundwater promotes this anaerobic reaction because it is deep underground and is covered with sand and rocks which results in little or no oxygen [4]. This reaction, however, is still able to be performed under aerobic conditions [1].
Salinity
Studies have shown that different levels of salinity affects which microorganisms are involved in the reduction of soluble U(VI). Some uranium contaminated groundwaters have a high salinity level due the high amount of acid used for processing uranium ores [5]. These elevated salinity groundwaters inhibit the growth of some U(VI)-reducing microorganisms [5].
pH and Presence of Carbonate
Some oxic areas of groundwaters have been found to contain UO22+ and UO2OH+ that forms a complex with carbonate [1]. These groundwaters have a neutral pH and high levels of carbonate concentrations, this leads to the formation of UO2(CO3)22− and UO2(CO3)34− which are stable [1]. Most dissimilatory metal-reducing bacteria are able to reduce these complexes; however, certain bacteria such as the Desulfotomaculum reducens are not able to reduce the uranium in the complex [1]. This can cause major problems in certain areas of uranium contaminated groundwaters if the bacteria present at those locations are not able to reduce the U(VI).
Microbial Processes
There are different methods of removing uranium for groundwaters such as pump-and-treat technologies and simple ground water flushing; however, both of these methods have been proven to be extremely ineffective [6]. Studies in laboratories have shown a simple and effective method by using dissimilatory metal-reducing bacteria to reduce U(VI). Two of the known major groups that are capable of this process are: dissimilatory Fe(III)-reducing microorganisms and sulphate-reducing microorganisms [7].
Dissimilatory Metal Reduction
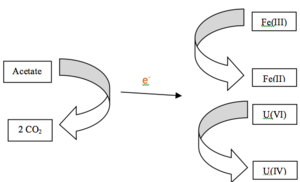
Geobacter metallireducens is known to be the most effective microorganism that is able to reduce U(VI) [8]. For this process to occur efficiently, reducible Fe(III) and acetate need to be added to the groundwater. Acetate acts as an organic electron donor to stimulate, enhance and promote anaerobic respiration which speeds up the reduction of U(VI) [6]. Reducible Fe(III) is essential for this process; without it, Geobacter will stop growing and the reduction of U(VI) will stop [8]. When both acetate and reducible Fe(III) is available, both U(VI) and Fe(III) are reduced to U(IV) and Fe(II) respectively [8]. In the end, Fe(II) and more importantly, insoluble U(IV), are seen to accumulate in the groundwater where the reduction process occurred [5]. This oxidation-reduction process keeps going until acetate and Fe(III) is used up.
Sulphate reducing microorganisms have been found to also reduce U(VI) both directly and indirectly [9]. Studies have shown that in contaminated groundwaters, sulphate reducing microorganisms indirectly reduces U(VI) by reducing sulphate to sulphide [1]. U(VI) is then rapidly reduced by sulphide. Furthermore, some sulphate reducing bacteria is also able to directly reduce U(VI) by oxidizing H2 and using U(VI) or Fe(III) as the electron acceptor instead of sulphate [1].
Key Organisms
Studies done on the uranium bioremediation in groundwaters have shown that microorganisms that are able to perform dissimilatory metal reduction are involved in the reduction of soluble U(VI) to insoluble U(IV). Fe(III)-reducing and sulphate-reducing bacteria are the main bacteria types that are able to perform this process effectively.
Fe(III)-reducing Bacteria
-Geobacter metallireducens[1]
-Shewanella putrefaciens[1]
Sulphate-reducing bacteria
-Desulfovibrio desulfuricans[1]
Current Research
Currently studies are being performed to learn more about the metabolic state of natural microbial communities in these uranium contaminated environments. The genome sequences of Geobacter is currently being heavily studied to identify more proteins that could explain its physiological state during uranium bioremediation. Having greater knowledge of the physiological state of Geobacter during uranium reduction could potentially lead to a more effective use of dissimilatory metal-reducing bacteria to solve the problem of uranium contaminated groundwaters. [10]
Additionally, studies have shown that Geobacter is not able to grow in high salinities uranium-contaminated groundwaters and the U(VI) reduction process is done by strains of Pseudomonas and Desulfosporosinus. These two types of bacteria are similar to Geobacter metallireducens in that they are able to also reduce Fe(III) which means they are a type of dissimilatory metal-reducing bacteria. This is extremely important especially in high salinities groundwaters where Geobacter is not able to grow. Currently, attempts are being made to isolate these salt-tolerant bacteria to study their physiology. [5]
References
[1] Wall J.D., Krumholz L.R. (2006) Uranium reduction. Ann Rev Microbiol 60:149–166.
[2] Hwang, C., Wu, W., Gentry, T.J., Carley, J., Corbin, G.A., Carroll, S.L., Fields, M.W. (2009). Bacterial Community Succession During in situ Uranium Bioremediation: Spatial Similarities Along Controlled Flow Paths. Journal Name: ISME Journal. doi: 10.1038/ismej.2008.123.
[3] Francis, A.J. (1998). Bioremediation of Uranium Contaminated Soils and Wastes. In Uranium Mining and Hydrogeology II, Freiberg (DE), 09/15/1998--09/17/1998.
[4] Jenkins, S. H. (1980). Water Treatment Handbook, Degremont: 1979. Fifth English Edition. FIRMIN-DIDOT S.A. PARIS, 1186 pp. Water Research, 14(1):93.
[5] Nevin K.P., Finneran K.T., Lovley D.R. (2003). Microorganisms associated with uranium bioremediation in a high-salinity subsurface sediment. Appl Environ Microbiol 69: 3672–3675
[6] Anderson R.T., Vrionis H.A., Ortiz-Bernad I., Resch C.T., Long P.E., Dayvault R., Karp K., Marutzky S., Metzler D.R., Peacock A., White D.C., Lowe M., Lovley D.R.. (2003). Stimulating the in situ activity of Geobacter species to remove uranium from the groundwater of a uranium- contaminated aquifer. Appl. Environ. Microbiol. 69: 5884 –5891.
[7] Holmes D.E., Finneran K.T., O’Neill R.A., Lovley D.R. (2002). Enrichment of members of the family Geobacteraceae associated with stimulation of dissimilatory metal reduction in uranium contaminated aquifer sediments. Appl. Environ. Microbiol. 68:2300 –2306.
[8] Barlett, M., Zhuang, K., Mahadevan, R., and Lovley, D. (2012). Integrative analysis of Geobacter spp. and sulfate-reducing bacteria during uranium bioremediation, Biogeosciences, 9, 1033-1040, doi: 10.5194/bg-9-1033-2012, 2012.
[9] Lovley, D. R., Roden, E. E., Phillips, E. J. P., & Woodward, J. C. (1993). Enzymatic iron and uranium reduction by sulfate-reducing bacteria. Marine Geology, 113(1–2), 41-53.
[10] Wilkins M.J., Verberkmoes N.C., Williams K.H., Callister S.J., Mouser P.J., Elifantz H., N’Guessan A.L., Thomas B.C., Nicora C.D., Shah M.B., Abraham P., Lipton M.S., Lovley D.R., Hettich R.L., Long P.E., Banfield J.F. (2009). Proteogenomic monitoring of Geobacter physiology during stimulated uranium bioremediation. Appl. Environ. Microbiol. 75:6591–6599.