Vibrio anguillarum
Classification
Higher order taxa
Domain: Bacteria
Phylum: Proteobacteria
Class: Gammaproteobacteria
Order: Vibrionales
Family: Vibrionaceae
Genus: Vibrio
Species
Vibrio anguillarum
Description and significance
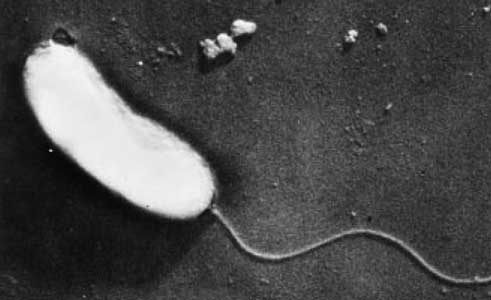
Vibrio anguillarum is a Gram-negative bacterium that belongs to the family Vibrionaceae, which includes several other species of marine bacteria. It is classified as a facultatively anaerobic, motile, and curved rod-shaped bacterium.
There are several serotypes of Vibrio anguillarum, which are distinguished based on differences in the structure of the bacterial surface antigens. These serotypes are important for understanding the epidemiology of the disease caused by V. anguillarum in aquatic animals, such as fish and shellfish.
Vibrio anguillarum causes vibriosis in abundance of aquatic species, more than 50 fresh and salt-water fish species, which includes salmon, Salmo salar and rainbow trout, Oncorhynchus mykiss (Frans et al., 2011).
16S Ribosomal RNA Gene Information
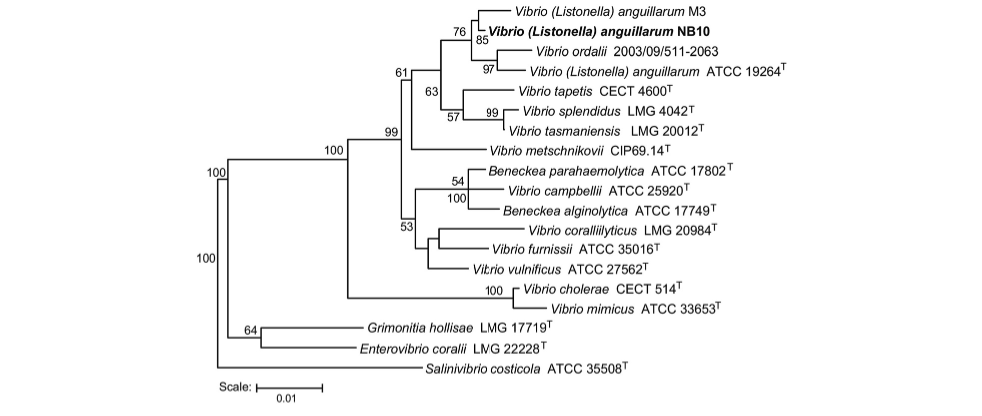
Vibrio anguillarum and Vibrio ordalli are the closest relatives according to microbiologists and those who have reported so to the NIH, in addition, the resemblance that the two species share has caused fervent debate and discourse on whether they truly are two separate species, as previously conducted identifications perhaps mistake V. ordalli for V. anguillarum and vice versa (Steinum et al., 2016).
Genome Structure
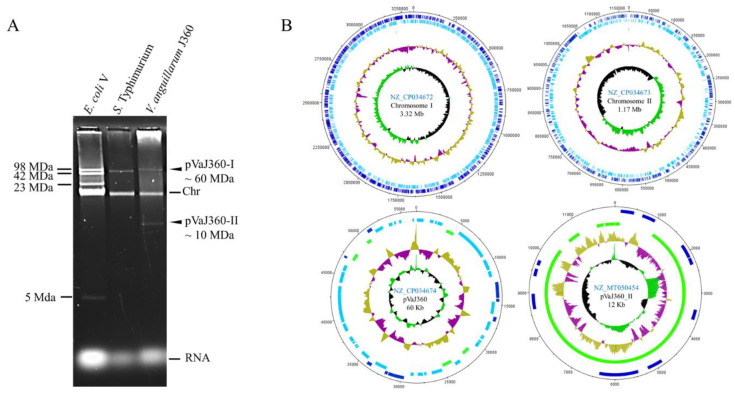
The total complete genome size of Vibrio anguillarum consists of 4,373,835 bp from the NB1O strain, as well as consisting of two circular chromosomes and one plasmid. V. anguillarum contains 3,783 protein-coding genes and 129 RNA genes. This sequencing was conducted within NSC(Nevada State College) in an effort to report the genome sequences of seven total V. anguillarum strains from multiple locations in an effort to examine its virulence and analyze genetic variations (Holm et al., 2018).
Cell structure and metabolism
The cell structure of Vibrio anguillarum consists of polar flagellum, a structure that is derived of four-five flagellin proteins, it is also covered in a membranous sheath.(Ormonde et al., 2000) Flagellin proteins are found in the hollow filament that makes the flagellum itself.
An intriguing function of the V. anguillarum is its behavior in the attachment and penetration of cells, in order to conduct its virulence in the form of the vibriosis that infects fish and crustaceans with. Some research suggests that V. anguillarum makes usage of an adhesin in order to properly attach to cells (Ormonde et al., 2000).
In terms of the metabolic functions that the bacterium exhibits, through expert analysis, it was discovered that V. anguillarum strain RV22 was able to adapt to conditions that consisted of low iron availability. It was discovered that the strain was able to lower the regulation of its energy during metabolism. These results showed that V. anguillarum was capable of adjusting its behavior in accordance to changes in its internal environment (Lages et. al., 2019).
Ecology and Pathogenesis
Vibrio anguillarum is pathogenic to an abundance of aquatic species, such as many mollusks, crustaceans and fish. Function akin to a hemorrhagic ailment, it infects the host with vibriosis. Specifically, it has been analyzed in infecting the chinook salmon species. Through the clinical abundance of signs that infect the host, which includes external symptoms of weight loss, lethargy, and noticeable red marks on the ventral and lateral areas of the fish. Swollen lesions, dark spots also form and ulcerate (Frans et al., 2011).
Current Research
Conducted by the Department of Microbiology and Parasitology within Spain, there has been research to further understand the role that Vibrio anguillarum plays as a virulent factor within the aquatic ecosystem. What was noted from this research, was that the bacterium optimally grew in temperatures around 25°C or 77 degrees fahrenheit. In addition, that the bacterium was capable in adapting and changing its behavior in accordance to its environment (Lages et al., 2019).
Specifically, the expression of its virulence factors differed in accordance to temperature such as the Chemotaxis, motility and even certain genes were expressed in differing temperatures than others. (Lages et al., 2019)
References
Frans, I, et al. “Vibrio Anguillarum as a Fish Pathogen: Virulence Factors, Diagnosis and Prevention.” Journal of Fish Diseases, vol. 34, no. 9, 2011, pp. 643–661., https://doi.org/10.1111/j.1365-2761.2011.01279.x.
Holm, Kåre Olav et al. “Complete genome sequence of Vibrio anguillarum strain NB10, a virulent isolate from the Gulf of Bothnia.” Standards in genomic sciences vol. 10 60. 2 Sep. 2015, doi:10.1186/s40793-015-0060-7
Lages, Marta A., et al. “The Expression of Virulence Factors in Vibrio Anguillarum Is Dually Regulated by Iron Levels and Temperature.” Frontiers, Frontiers, 25 Sept. 2019, https://www.frontiersin.org/articles/10.3389/fmicb.2019.02335/full.
Ormonde, P et al. “Role of motility in adherence to and invasion of a fish cell line by Vibrio anguillarum.” Journal of bacteriology vol. 182,8 (2000): 2326-8. doi:10.1128/JB.182.8.2326-2328.2000
Steinum, Terje M et al. “Multilocus Sequence Analysis of Close Relatives Vibrio anguillarum and Vibrio ordalii.” Applied and environmental microbiology vol. 82,18 5496-504. 30 Aug. 2016, doi:10.1128/AEM.00620-16
Vasquez, Ignacio et al. “Comparative Genomics Analysis of Vibrio anguillarum Isolated from Lumpfish (Cyclopterus lumpus) in Newfoundland Reveal Novel Chromosomal Organizations.” Microorganisms vol. 8,11 1666. 27 Oct. 2020, doi:10.3390/microorganisms8111666
Author
Created by Zachary Perez, a student of Dr. Hidetoshi Urakawa of Florida Gulf Coast University for Microbial Ecology EVR 4024C spring 2023.