Brucella microti
A Microbial Biorealm page on the genus Brucella microti
Classification
Higher order taxa
Bacteria (Domain); Proteobacteria (Phylum); Alpha Proteobacteria (Class); Rhizobiales (Order); Brucellaceae (Family)
Species
Brucella microti
NCBI: Taxonomy
Description and significance
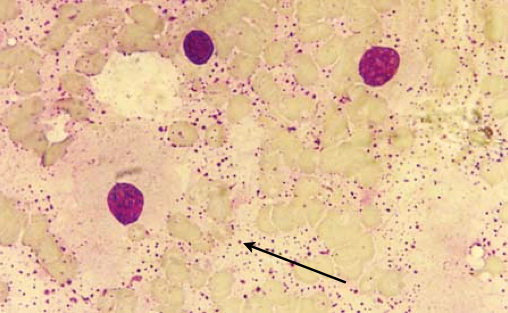
B. microti was first isolated from the liver of the common vole (Microtus arvalis) found in South Moravia, Czech Republic in 2000[2]. Figure 1 shows the presence of this species in the blood from the heart of a vole. The cells are non-motile, gram negative cells that have a slightly elongated spherical shape[2]. The phenotype of B. microti strongly resembles Ochrobactrum, which is a genus of known human pathogens, more closely than it resembles Brucella[3]. Because of its similarity to the Ochrobactrum genus, this species was originally misidentified as a new species in the genus Ochrobactrum[4]. More recently, B. microti was also identified from red foxes from Lower Austria[3]. The frequent isolations from different mammalian hosts in different geographical regions suggests that B. microti could be an emerging pathogen[3]. Most recently, B. microti was isolated from the soil where the infected voles were found, which is atypical for Brucella species[5]. The persistence of this bacteria outside of a mammalian host further suggests that this species could be pathogenic and potentially lead to the reemergence of brucellosis, a worldwide zoonotic infectious disease[4,5]. Brucellosis causes malta fever in humans, which is characterized by fever, severe fatigue, muscular pain, and sweating. If left untreated, brucellosis can develop into a chronic infection and lead to endocarditis, osteoarthritis, and neurological damage[6]. Brucellosis is also prevalent in animals and can cause sterility and abortion[6].
Genome structure
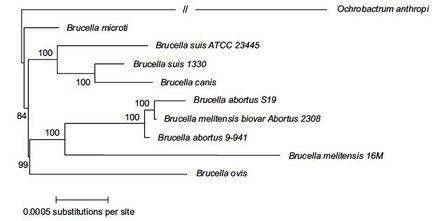
The genome of B. microti was sequenced in 2009. It was found that the genome consists of two separate circular chromosomes; one 2,117,050 base-pairs long and the second 1,220,319 base-pairs long. From this sequencing, 3,291 protein coding genes, 55 tRNA’s, and 9 ribosomal RNA’s have been predicted for this species[3]. It was also determined that the genome of B. microti is almost identical to B. suis, a known human pathogen. The sequence of B. microti was 99.84% identical to B. suis in aligned regions, suggesting that these two species are close genetic relatives[3]. Within the genus Brucella, DNA sequence analysis revealed that B. microti forms a separate clade in the genus but is still closely related to B. suis[7].
One notable difference that separates B. microti from the other Brucella species is that B. microti has a unique variation in its 23S ribosomal DNA sequence, which could be the reason why this species is capable of increased growth compared to other Brucella species[3,7]. This variation appears to be caused by the cleavage of the intervening sequences of the 23S rRNA by RNase III during the maturation process. This alternative cleavage results in a specific fragmentation pattern of the 23S rRNA that is unique from other Brucella species, with the exception of B. inopinata, which also has this fragmentation pattern and has a faster growth rate than the typical Brucella species[3]. Also, four functional genes were found in B. microti that are impaired in all other Brucella species. The first gene produces an enzyme involved in pyruvate metabolism. The second catalyzes a reaction that produces succinate, which is involved in many metabolic pathways and is used in physiological reactions. The third gene produces a protein component of an ABC transporter and is involved in dipeptide transport. And the forth encodes for transcriptional regulators that mediate the cellular response to environmental stress and resistance to antibiotics[3]. Interestingly, these four genes are also active in Ochrobacterium anthropi, which is a close relative and phenotypically similar to B. microti[3].
Cell and colony structure
B. microti cells are gram-negative coccobacilli, or slightly elongated coccus cells, 0.5-0.8µm in diameter and 0.6-1.4 µm in length[2]. Cells could be found both individually and arranged in irregular clusters. The cells are unflagellated, non-motile, and lack the capability to form spores[2].
B. microti has an optimum growth temperature of 37℃ but is also able to grow in temperatures ranging from 25℃ to 42℃[2]. This species has been cultured on a variety of media including meat peptone agar (MPA), trypticase soy agar (TSA), blood agar, standard nutrient agar, and Brucella agar without supplementary CO2[2,4]. After 1-2 days of incubation, the colonies grew to a diameter of 1-2 mm and were a transparent to whitish color[2]. Colonies appeared to be smooth and slightly concave with continuous edges. After 72 hours of growth, the colonies grew to 6-9 mm in diameter and most had developed a brown coloring[4].
Metabolism
B. microti has exhibited outstanding metabolic capabilities compared to all other Brucella species[2]. It is a fast growing aerobic bacterial species and is much more biochemically active than other Brucella species[3], with a generation time of 2 hours, compared to the 3 hour generation time of B. suis[8]. It is a facultative intracellular species and it has been observed to be non-fastidious[4]. B. microti tested positive for oxidase, catalase, and urease. The cells are also capable of reducing nitrate and nitrite, can produce nitrogen gas from nitrate, and are not able to produce H2S gas[2].
Ecology
B. microti has adapted to specific mammalian hosts, two of which have been identified: the common vole and the red fox. This species has developed acid resistant adaptations, such as the glutamic acid decarboxylase (GAD) system, that functions to overcome the low pH of the gastrointestinal tract of their host[3]. In 2007, B. microti was successfully isolated from the soil at the same site as the first isolation from the common voles in 2000[5]. Because 7 years had elapsed and living cells were still successfully isolated, this species has demonstrated its capability of long-term survival in the soil. The species could also still be isolated from the soil samples after being kept in storage at 4℃ for 6 months[5]. The capability to survive for long periods of time outside of a mammalian host in soil could be a potential primary reservoir for infection, which could facilitate the reemergence of brucellosis[4,5].
Pathology
The pathogenicity of B. microti in humans and livestock has not yet been confirmed[4], however, this new species has exhibited a high pathogenic potential in contrast to other Brucella species[6,8]. By studying B. microti infections in mice, it was found that this species was highly pathogenic for mice due to an increased ability to replicate, which suggests that B. microti is an emerging pathogen[9].
In comparison to B. suis, a known human pathogen, B. microti showed faster growth and an increased acid resistance upon infection of both BALB/c and C57BL/6 mice. After 24 hours past infection, the number of intracellular B. microti cells present were about 100 times greater than the number of intracellular B. suis cells[8]. Within 4 to 7 days after infection, B. microti killed 82% of the infected mice. Also, the peak of infection in the blood, liver, and spleen of mice infected with B. microti was reached 4 days sooner than the peak of infection in the mice infected with B. suis[8]. This increased rate of replication and severe infection in mice suggests that B. microti could be a threatening human pathogen and needs to be studied further[8].
It has been observed that the immediate acidic environment around the cells actually triggers the intracellular replication of B. microti[6]. Also, a key virulence factor of B. microti is the use of the Type IV Secretion System (T4SS) which is encoded by the virB operon. This operon is induced only when the cell is in acidic environments, such as the gastrointestinal tract of their host[6]. It has been shown that the virB operon is essential for the ability to reproduce in such environments, and that B. microti cannot survive or create a persistent infection without this operon being present[6].
The glutamic acid decarboxylase (GAD) system, which is used by Escherichia coli and other acid resistant bacteria, is known to contribute to the survival of bacteria cells in extremely low pH environments. The system works by the use of an antiporter to bring glutamate into the cell while exporting gamma-aminobutyric acid (GABA) into the extracellular medium, resulting in an elevated pH in the extracellular environment[10]. It was recently discovered that the GAD system was also used by B. microti to resist acidic stress, in contrast to B. suis, which does not have the GAD system. Even at a pH of 2.5, 20% of B. microti remained viable in contrast to complete death in all B. suis cells, suggesting that the GAD system plays a significant role in the survival and reproduction of B. microti in pH values characteristic of the gastrointestinal tract[10].
References
[1] Centre of Activity of the ASCR. <http://www.cas.cz/o_avcr/zakladni_informace/dokumenty/anotace/2007/6_sekce_biologicko_ekologickych_ved.html> Accessed Mar, 13, 2013.
[2] Scholz, Holger, Z Hubalek, I Sedlacek, et al (2008). Brucella microti sp. nov., isolated from the common vole Microtus arvalis. International Journal of Systematic and Evolutionary Microbiology. 58: 375-382; doi:10.1099/ijs.0.65356-0.
[3] Audic, S, M Lescot, JM Claverie, and H Scholz (2009). Brucella microti: the genome sequence of an emerging pathogen. BMC Genomics. 10: 352-369; doi:10.1186/1471-2164-10-352.
[4] Al Dahouk, Sascha, E Hofer, H Tomaso, et al (2012). Intraspecies Biodiversity of the Genetically Homologous Species Brucella microti. Applied and Environmental Microbiology. 78(5): 1534-1543; doi:10.1128/AEM.06351-11.
[5] Scholz, Holger, Z Hubalek, J Nesvadbova, et al (2008). Isolation of Brucella microti from Soil. Emerging Infectious Diseases. 14(8): 1316-1317; doi:10.3201/eid1408.080286.
[6] Hanna, Nabil, M Jimenez de Bagues, S Ouahrani-Bettache, et al (2011). The virB Operon is Essential for Lethality of Brucella microti in the Balb/c Murine Model of Infection. Journal of Infectious Diseases. 203: 1129-1135; doi:10.1093/infdis/jiq163.
[7] Ficht, Thomas (2010). Brucella taxonomy and evolution. Future Microbiology. 5(6): 859-866; doi:10.2217/fmb.10.52.
[8] Jimenez de Bagues, Maria, S Ouahrani-Bettache, J Quintana, et al (2010). The New Species Brucella microti Replicates in Macrophages and Causes Death in Murine Models of Infection. Journal of Infectious Diseases. 202: 3-10; doi: 10.1086/653084.
[9] Jimenez de Bagues, Maria, A Martino, J Quintana, et al (2011). Course of Infection with the Emergent Pathogen Brucella microti in Immunocompromised Mice. Infection and Immunity. 79: 3934-3939; doi:10.1128/IAI.05542-11.
[10] Occhialini, Alessandra, M Jimenez de Bagues, B Saadeh, et al (2012). The Glutamic Acid Decarboxylase System of the New Species Brucella microti Contributes to its Acid Resistance and to Oral Infection of Mice. Journal of Infectious Diseases. 206: 1424-1432; doi: 10.1093/infdis/jis522.
Edited by Kelsey Toncer of Dr. Lisa R. Moore, University of Southern Maine, Department of Biological Sciences, http://www.usm.maine.edu/bio