Porcine reproductive respiratory syndrome virus
Introduction
By Rhea Le
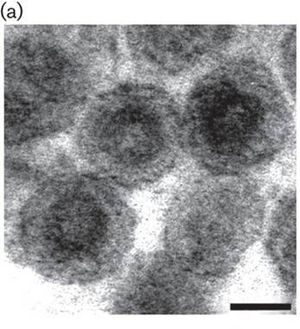
The porcine reproductive and respiratory syndrome virus (PRRSV) is from the Arteriviruses and Nidovirus order ( Fig. 1 ). This family contains positive stranded RNA that infects mammals [11]. PRRS was first reported in the United States in 1987 and was then isolated and classified in the early1990s. Interestingly, swine macrophages are the only cell type to be found to support PRRSV replication. PRRSV causes porcine reproductive and respiratory syndrome, which can cause high mortality rates among swine population [1]. This disease causes serious economic losses to swine-producing countries worldwide [5]. More specifically, PRRSV causes late-term reproductive failure and pneumonia in infant pigs [6]. PRRSV inflicts damage upon swine by attacking healthy macrophages, especially ones located in the lungs or alveolar macrophages. There have not been many super effective vaccines for the virus due to the highly variable effects seen in infected swine. Therefore, it is crucial for us to learn about these viruses and investigate how these viruses infect their hosts to possibly produce more effective vaccines and control the virus infection.
PRRSV Structure and Genome
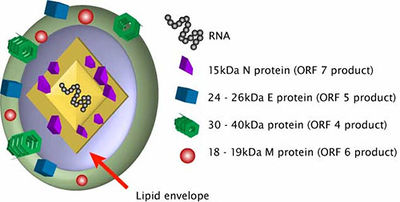
PRRSV is a part of the Arterividae family. The genomes from this viral family are about 12 to 16 kb long with the 3’-poly(A) tail and the 5’ cap. There are 10 to 15 known ORFs. ORF1a is within the 5’-proximal region of the genome and is responsible for encoding the gene for polyprotein 1a. ORF1b is located in the same region and is expressed in ribosomal frameshifting. There also exists a short trans frame (TF) ORF that overlap the ORF1a and is expressed by the -2 PRF. This TF ORF is conserved in viruses such as PRRSV, elevating virus (LDV), and simian hemorrhagic fever virus (SHFV), but not equine arteritis virus (EAV). ORFs in the 3’-proximal region are code for structural proteins (Fig. 2). The arterivirus genome is contained inside a single N protein of a core with about 110-128 amino acids. The nucleocapsid has been thought to be isometric but it actually might be a pleomorphic core structure with a mean diameter of 39 nm. The N-protein is phosphorylated and its terminal site has positively-charged residues that can bind to RNA [11].
The arterivirus genome is contained inside a single N protein of a core with about 110-128 amino acids. The nucleocapsid has been thought to be isometric but it actually might be a pleomorphic core structure with a mean diameter of 39 nm. The N-protein is phosphorylated and its terminal site has positively-charged residues that can bind to RNA or be involved in its interactions with other proteins. The virions have a sucrose density of 1.13 to 1.17 g cm-1 and the envelope is quite smooth. Scientists believe that the arterivirus envelope contains two major and five minor proteins. PRRSV has the GP5 protein and the M protein. Other arteriviruses have the major GP protein instead of the GP5 protein. The GP5 and M protein forms a heterodimer that controls viral assembly and the transportation of the proteins to the Golgi apparatus. This step is important for host infection. These proteins form a disulfide-linked heterodimer that is required for virus assembly. Other important proteins are the minor proteins GP2, GP3, and GP4. These minor proteins also form disulfide-linked heterodimers. Lastly, there are two minor non-glycosated polypeptides (proteins) that are located within the viral membrane. Especially with PRRSV, the small E protein is a minor structural protein that is essential for viral infection. The other small protein is hydrophobic and its functions are not currently clear [11].
PRRSV has a nucleocapsid with an asymmetric and linear arrangement [4]. Once it infects the cells, PRRSV can be identified under the microscope as interstitial thickening of macrophages and cell debris in the alveoli [10]. The viral genome is about 15 kb and consists of seven ORFs. The PRRSV replicase gene is composed of two large ORFs, ORF1a and ORF1b. The two ORFs make up about 80% of the entire genome. ORF1b is expressed in ribosomal frameshifting. ORFs 2 through 7 encode structural proteins. More specifically, ORFs 3, 4, 5 and 6 code for the virion proteins on the outer surface while ORF 7 code for the nucleocapsid proteins (Fig.2).
Different Strains of PRRSV
There are significant variations between the two main strains of PRRSV, VR-2332 and Lelystad. The amino acid sequences for these two strains are about 40% different from one another. The VR-2332 strain is the only one that exists in North America. While in Europe, both VR-2332 and Lelystad strains exist, with the Lelystad being the dominant strain. PRRSV exhibits a rate of mutation of 10-2 per amino acid sites per year as opposed to 10-3 to 10-5/ site/year for other RNA viruses. Scientists have suspected that PRRSV was transmitted to swine from another host in about 1980 because the estimated divergence time between these two strains is about 1972 to 1988 with an evolutionary rate of (4.71-9.8) x 10-2 [6]. This high rate of mutation is due to a faster rate of replication. Management factors can increase the rate of replication. One example is the dense population of swine, which allows PRRSV to be easily spread through the air from one pig to another. Moreover, it makes sense that swine at larger farms are more likely to be infected than smaller farms since larger farms are more likely to have a higher density of pigs. Also, since the virus is a relatively new pathogen in swine, pigs have not yet developed resistance against the virus. Currently, the influences of the host in regard to the breed or family of the swine are not known. However, infection of swine host with PRRSV and another pathogen, such as Mycoplasma hyopneumoniae or Strep. Suis, at the same time leads to a worse PRRSV infection. Thus, respiratory disease in infant swine is due to PRRSV but respiratory disease in grown swine could be due to PRRSV and M. hyopneumoniae.
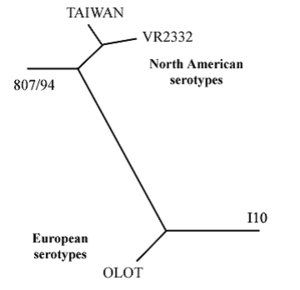
VR-2332 and Lelystad strains both exist in South America and Asia. Other prevalent strains include the strain originating from Taiwan, 807/94 from Canada, Olot from Spain, and I10 from The Netherlands. The percentage of homology between the VR-2332 strain and the Taiwan strain is 97%. The percentages of homology between VR-2332 and 807/94, Olot, and I10 are 92%, 66%, and 66%, respectively. A phylogenetic tree of 5 PRRSV strains can be seen in Figure 3.
Within the different strains of PRRSV, some exhibit more variation than others. The American strains that were isolated from infected swine showed more genetic diversity than the European strains. Sequences from the European isolates were more conserved and related to one another. Strains from The Netherlands, Spain, France and Great Britain had a homology of 96-100% for the ORF7 gene and 98-100% homology for the sequence that codes for the nucleocapsid protein [2].
In contrast, a strain found in Belarus (Belarusian EU-PRRSV) was found to have the genotypes from the European strains and contain high levels of diversity. This diversity is marked by size polymorphism in ORF7 from 375 to 393 nucleotides. Also, Belarusian EU-PRRSV contains more diversity than North American genotypes. This high level of diversity, also seen in other European strains east of the Polish border, is likely due livestock trading between Belarus and the west. The ancestral populations from the west, specifically North America, contain more genetic variation that the descendant population in Europe [12].
PRRSV Pathogenesis
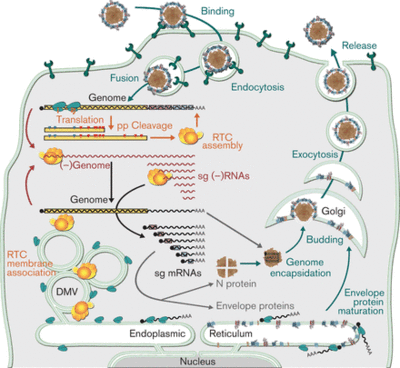
Macrophages are phagocytic white blood cells that make up the first step in cellular defense against pathogens [10]. The virus enters the cell via clathrin-mediated endocytosis where the viral membrane fuses with the endomal membrane to release the viral ssRNA into the cell. The host membrane has heparin-like molecules that are believed to interact with the sialic acids on the surface of the virion. Sialic acids have been shown to be important for infection. Endocytosis is mediated by sioadhesin (CD169), which an immunoglubin-like lectins (carbohydrate-binding proteins) that carry sialic acids. Additionally, the GP5/M heterodimer interacts with the sialic acids [3]. Once in the cytoplasm, it translates the replicase polyproteins 1a and 1ab. These polyproteins are then cleaved by internal proteinases and interact with non-structural proteins to assemble a replication and transcription complex (RTC). This complex produces a full-length genome and a minus-RNA strand. The minus-RNA strand is used as a template for the synthesis of sg mRNA, which expresses structural protein genes. The replication takes place in viral factories, which increase the efficiency of viral replication. The new genomes are assembled into the nucleocapsids at the host endoplasmic reticulum (ER) and then released via exocytosis.
The virus replicates in the perinuclear cytoplasm of the host macrophages. And the virions are released via exocytosis to the outside of the cell. Also, it reduces the production of pro-inflammitory protein, TNF-α and IFN-α. These proteins are important because they induce host resistance to viral replication through the destruction of viral mRNA and viral gene translation. Also, theses proteins activate NK cells that selectively kill viruses. The overall effect of in this step is a poor intracellular response to the virus. Furthermore, PRRSV decreases the rate of phagocytosis and causes apoptosis. A full depiction of the PRRSV infectious cycle within the host cell can be seen on the right (Fig. 3).
When arteriviruses infect host cell, the double membrane vesicles (DMVs) interact with the viral replication and transcription complex (RTC). DMVs are believed to be formed by nnsp2, nsp3, and nsp5. Furthermore, cyclophilin A is believed to be involved in arterivirus RNA synthesis from results of inhibitor studies with cyclosporine A drug. Cyclopphilin proteins are found in vertebrates and act as immunosuppressants, which are commonly used to suppress a patients immune system after transplant [13]. Also, studies suggest that arterivirus replication is promoted by autophagy. Accordingly, PRRSV N proteins were found in the nucleoli of infected host cells. This could be due to the interactions between nuclear host proteins, such as fibrillarin, nucleolin, and poly(A)-binding proteins, and PRRSV N proteins [11].
IFN-β production in the host cell is modified by the NendoU endoribonuclease, nsp1-α and nsp1-β. The NendoU enzyme is known to be highly cytotoxic when it is expressed. PRRSV nsp1-α and nsp1-β localize at the host nucleus and suppress the expression of IFN-β. More specifically, nsp1-α suppresses NFxB activation and nsp1-β inhibits IFN regulatory factor 3 (IRF3) production. Also, nsp1-β inhibits downstream IFN signalling pathways via the STAT1 phosphorylation and ISGF3 translocation. Recent research has shown that a conserved nsp1-β motif is involved in inhibiting the IFN response [4].
PRRSV nsp1α, nsp1β, nsp2 and nsp7 all induced an immune response, with the presence of specific antibodies being seen from 14 to 202 days after infection. Also, neutralizing antibodies are produced in PRRSV- infected animals pretty late and in low concentrations. Furthermore, in regard to PRRSV, the expression of hypoglycosated GP5 induce higher levels of neutralizing antibodies in infected swine. This effect could be due to an immune-dominant epitope in GP5. The late presence of and low concentration of neytraling antibodies is believed to be important for PRRSV infection because it prevents the development of normal B cells. In addition, there have been reports of cell-mediated immune response to PRRSV infection. However, the specific T cell concentrations in IFN-y-producing cells are highly variable in swine such that there is no correlation with host antibody concentration and viral concentration [11].
Additionally, sialic acids that carry a complex of N-linked glycans are associated with PRRSV infection. The attachment of PRRSV to the porcine alveolar macrophages somewhat depends on the presence of sialic acids on the exterior surface of the virus. Some PRRSV structural proteins that might contain N-linked glycans and sialic acids are GP2a, GP3, GP4, and GP5. Neutralizing epitopes have been shown to be involved in viral interactions with host receptors in GP4 and GP5. More specifically, a highly conserved N-linked glycosation site used to attach to receptors is seen in GP5 proteins of VR-2332 and Lelystad PRRSV strains. However, a monoclonal antibody that neutralizes epitopes of both strains has been found. α -2-9 linkages, types of lectins, have are not known to be involved in PRRSV infection but recently. However, data has shown that α -2-3 and α -206 are involved in PRRSV infection [3].
Some clinical reproductive problems of PRRSV infection include late gestation and early gestation problems. Early gestation symptoms include abortions or absorption of fetus. Late gestation problems can be seen with the presences of weak, stillborn and mummies in litters. Moreover, respiratory problems are usually associated with the presence of pneumonia. PRRSV can enter the pig via inhalation, ingestation, vaccination, insemination, and other pathways. In the case of insemination, the virus travels throughout the body of the infected male boar via circulation to the semen. Once the semen is in the uterus, the virus replicates in the host macrophages, which leads to regional lymph nodes and appearance of viremia. During conception, the virus will replicate in the fetal lymphoid tissues, which includes the thymus, tonsils, and lymph nodes. Within herds, however, the effects of the virus are unpredictable since some pigs suffer PRRS while others show no symptoms [8].
Replication, Transcription, and Translation
A conformation at the 3’proximal RNA hairpin acts as a pseudoknot interaction with a hairpin located upstream within the N protein gene is conserved in all arteriviruses. This conformation may involved in the negative-strand RNA synthesis. Another conserved region occurs at the 3’-terminal chemokine (C-C) motif, which is located upstream from the poly-(A) tail. This motif could be important for replication and/or negative-strand RNA synthesis. Additionally, a kissing stem-loop, which is formed when two bases between two haipin loops in RNA pair up with one another [14]. The two hairpin loops involved in this interaction are located at the 3’temrinal of NTR and the N protein. This interaction is thought to play a major role PRRSV replication.
The translation of arterivirus genomes is believed to initiate with ribosomal scanning of the 5’NTR and include a few framshifts. The framseshift efficiency is estimated to be about 15-20%. Most arteriviruses (except EAV) use -2 PRF to express a conserved TF ORF that is located in the doing region of nsp2. This site codes for three products, nsp2, nsp2TF, and nsp2N. In PRRSV, about 20 % of ribosomes that translate the nsp2 region translate nsp2TF, about 7% translate nsp2N and about 15% translate ORF1b [9].
Post-Translational Processes
The post-translational processing of replicase polyproteins is directed by four ORF1a domains for PRRSV and LDV and is directed by three ORF1a domains for EAV. The cleavage of the pp1a and pp1b produces 14 end products for PRRSV, namely nsp1 to 12. For PRRSV, LDV, and SHFV, the -2 PRF creates truncated nsp2 products called nsp2TF and nsp2N. Alternative cleavage can also occur in the nsp3-8 region.
All arteiviruses have nsp 1, 2, and 4 that contain two functional papain-like proteinases (PLPs) and one functional serine proteinase (SP). Of PLP1- α and PLP1-β, the latter has been inactivated in EAV, Furthermore, PRRSV and LDV contain another non-structural proteinase that is involved in the cleavage of nsp1 to nsp1- α and nsp1-β. The two PLPs could be the result of a duplication event. There exists four proteinase domains in arteivirusse, with PLP-α and PLP-β being very compact domians that contain Cys-His tandem active-site residues. Aside from the PLP1, the PLP2 domain has cis and trans cleavage properties. Also, it is thought to be a subclass of zinc-dependant ovarian tumor domain-containing deubiquitinases (OUT DUBs) due to the presence of a zinc finger in its domain. Lastly, the crystal structures of nsp4 in EAV and PRRSV indicate two domains, which form the two- β-barrel fold, and a C-terminal domain that is involved in replicase polyprotein processing.
After the cleavage of pp1a and pp1b, the non-structural protein products assemble to an enzyme complex that directs replication and transcription. For the nsp2 proteins, nsp2TF consists of an alternative transmembrane protein that is involved in the exocytosis and the nsp2N is truncated and lacks a hydrophobic domain. In cells that are infected with PRRSV, scientists have found additional nsp2 and nsp2TF proteins. In addition, the nsp2 protein has a hyper-variable region, where deletions and insertions have been found for PRRSV and EAV. Although the activities of this region are not clear, the variation might be important to the virulence of the virus, especially for the Asian strains of PRRSV. This could be a key research area for future studies of the virus.
Arteriviruses contain RNA-dependent RNA polymerase and helicase domain in the ORF1b that share a common ancestry with coronaviruses. Also, nsp10 contains the N-terminal zinc-binding domain that is conserved in all families of the Nidovirus domain. Another conserved ORF1b domain codes for an endoribonuclease function, NendoU (nidovirus endocuclease specific for uridylate). The functions of NendoU in the virus and the host cells are not currently known. Two other ORF1b domains, the third N-terminal of nsp9 and the C-teminal nsp-12 product, have not been characterized.
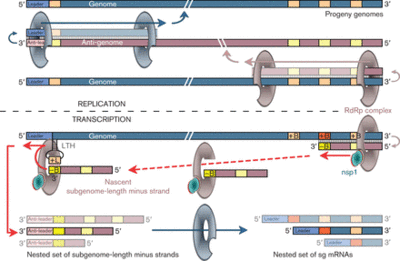
The translation of the upstream ORF ends in the dissociation of the ribosome from the RNA at the stop codon so that the ORF downstream would not be translated. Subgenomic mRNA or sg mRNA is a special type of RNA used by viruses to translate an ORF downstream of another ORF [13]. There is a subgenomic promoter sequence on the negative RNA strand that is recognized by an RNA dependent RNA polymerase (RdRp) to generate a short sequence that contains the sequence necessary for the translation of the downstream ORF, this is sg mRNA. A distinct synthesis of sg-mRNAs is seen in only Arteriviruses and coronaviruses of the Nidovirus order. Arterivirus and coronavirus genomes contain identical 5’ ends and a “leader sequence” (about 156-211 nucleotides long) and 5’-proximal regions. Discontinuous RNa synthesis could be responsible for generating discontinuous negative-strand RNAs, which then serve as templates for sg mRNA synthesis. Transcription of sg mRNAs depend on a transcriptional regulatory sequence (TRS) duplex formation. Additionally, sg mRNAs are produced non-equimolar and are structurally polycistronic and functionally monocistronic [11]. The entire synthesis of sg mRNA can be viewed in Figure 5.
PRRS Treatment
Modified live virus (MLV) and inactivated vaccines are used to treat arteriviruses. PRRSV MLV was available 1994 to the US and Europe and have been able of control disease outbreaks. But these vaccines are not efficient for any viral mutations since they were developed from a single strain. Also, MLV can undergo recombination with field isolates such that their protection against PRRSV had been questioned. Another problem is that the PRRSV infection is very difficult to predict in swine since the virus shows much variation within infected swine. Therefore, vaccines for PRRSV are difficult for scientists to develop. However, plasmid DNA and viral vector vaccines that contain PRRSV structural proteins have been developed. But MLV vaccines are still the preferred method of vaccination.
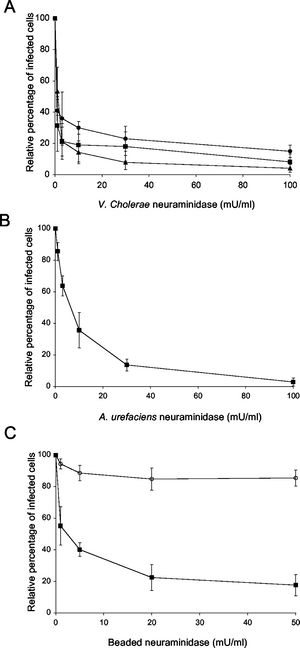
One study examined on the attachment of PRRSV to porcine alveolar macrophages by focusing on sialic acids. Since PRRSV uses sialic acids on its surface to bing to host cells, disrupting this event would decrease infection. The enzyme that the experimenters used is neuraminidase, which has a high preference for sialic acids and binds to them. Neuraminidase binding causes the sialic acids to not bind or not bnind tightly to the host cell. Neuraminidases from different sources were obtained so that their effectiveness could be compared. Figure 6 shows the effects of treating PRRSV-infected cells to neuraminidase. (A) shows the effects of V.cholare neuraminidase on infected cells; (B) shows effectiveness of neuraminidase from A. urefaciens; (C) shows the effectiveness of beaded neuraminidase [3]. As we can see, all types of neuraminidases had reduced infection significantly. However, V. cholerae neuraminidase was the most effective at reducing infection. Furthermore, results of this study can be used to produced new drugs or vaccines for PRRS.
To control PRRS, caretakers attempt to “stabilize” the infection in the herd by trying to achieve immunity in all sows. Of course, the male semen must also be free of PRRSV. This method ensures that all offspring will not inherit the virus and overtime, some sow herds that only produce uninfected offspring can be used to eliminate PRRS in the herd. The virus is known for its persistence in swine, with long-term carrier pigs. Fortunately, most infected swine has been recorded to gain immunity after about 60 days after being infected [8].
References
4. Dokland, T. (2010). The structural biology of PRRSV. Virus Research, 154(1-2), 86-97.
14. Nowakowski, J. & Tinoco, I., Jr. (1997)Semin. Virology.8,153–165.
