Halorhodospira halophila: Difference between revisions
No edit summary |
No edit summary |
||
| Line 44: | Line 44: | ||
''Halorhodospira halophila'' (formerly ''Ectothiorhodospira halophila'') is an extremely halophilic purple bacterium that was formally a member of the Ectothiorhodospira genus until recently reclassified. Phylogenetically, ''Halorhodospira halohila'' is associated within the gamma subdivision of the phylum Proteobacteria and is known to be phototrophic and Gram-negative. It is considered to be “one of the most halophilic eubacteria known.” [2] Its halophilic nature allows for it to be present in conditions that often have been thought to be too harsh for bacteria, and it is found proliferating in saturated salts such as crystallizer ponds (a hypersaline environment where sodium chloride precipitates) that have a salinity of 25% or higher. This importantly shows that harsh environments once thought to be exclusive to archaea actually contain bacteria as well. [3] | ''Halorhodospira halophila'' (formerly ''Ectothiorhodospira halophila'') is an extremely halophilic purple bacterium that was formally a member of the Ectothiorhodospira genus until recently reclassified. Phylogenetically, ''Halorhodospira halohila'' is associated within the gamma subdivision of the phylum Proteobacteria and is known to be phototrophic and Gram-negative. It is considered to be “one of the most halophilic eubacteria known.” [2] Its halophilic nature allows for it to be present in conditions that often have been thought to be too harsh for bacteria, and it is found proliferating in saturated salts such as crystallizer ponds (a hypersaline environment where sodium chloride precipitates) that have a salinity of 25% or higher. This importantly shows that harsh environments once thought to be exclusive to archaea actually contain bacteria as well. [3] | ||
This halophilic organism was first isolated from a salt lake mud [5] and has been determined to have several functions: production of organic solutes glycine betaine, ectoine, and trehalose to help balance osmotic pressure, as well as the oxidization of sulfide to sulfur (which later would be further oxidized into sulfate). [2] This has resulted in the use of ''Halorhodospira halophila'' for research on topics from | This halophilic organism was first isolated from a salt lake mud [5] and has been determined to have several functions: production of organic solutes glycine betaine, ectoine, and trehalose to help balance osmotic pressure, as well as the oxidization of sulfide to sulfur (which later would be further oxidized into sulfate). [2] This has resulted in the use of ''Halorhodospira halophila'' for research on topics from photobiological ability to produce hydrogen gas to effects on sulfur. | ||
One important aspect is the production of the bacterial photoreceptor PYP (photoactive yellow protein) within ''Halorhodospira halophila''. PYP is a blue-light sensor found in ''Halorhodospira halophila'' that has great significance towards protein research and biotechnology. Photoactive proteins are generally accepted as "model systems for studying protein signal state formation." As a small protein, PYP provides for an 'attractive model system for exploring how a chromophore and protein interact to sense light and send a biological signal." [4] | One important aspect is the production of the bacterial photoreceptor PYP (photoactive yellow protein) within ''Halorhodospira halophila''. PYP is a blue-light sensor found in ''Halorhodospira halophila'' that has great significance towards protein research and biotechnology. Photoactive proteins are generally accepted as "model systems for studying protein signal state formation." As a small protein, PYP provides for an 'attractive model system for exploring how a chromophore and protein interact to sense light and send a biological signal." [4] | ||
| Line 60: | Line 60: | ||
''Halorhodospira halophila'' is capable of a large number of metabolic pathways such as glycolysis, the citrate cylce, amino acid metabolism, and more. A full list of these pathways and their individual maps can be found at the [http://www.genome.jp/dbget-bin/www_bfind_sub?mode=bfind&max_hit=1000&dbkey=kegg&keywords=halorhodospira+halophila KEGG Pathway Database for Halorhodospira halophila]. [8] | ''Halorhodospira halophila'' is capable of a large number of metabolic pathways such as glycolysis, the citrate cylce, amino acid metabolism, and more. A full list of these pathways and their individual maps can be found at the [http://www.genome.jp/dbget-bin/www_bfind_sub?mode=bfind&max_hit=1000&dbkey=kegg&keywords=halorhodospira+halophila KEGG Pathway Database for Halorhodospira halophila]. [8] | ||
One important part of <I>Halorhodospira halophila</I> (and subject to recent research) is the | One important part of <I>Halorhodospira halophila</I> (and subject to recent research) is the presence of Photoactive Yellow Protein (PYP), a 14kDa cytoplasmic photoreceptor protein, which is an important protein for current research. | ||
''Halorhodospira halophila'' also has a potential role of hydrogen production through the nitrogen fixation of nitrogenase, which will be further explained below. | ''Halorhodospira halophila'' also has a potential role of hydrogen production through the nitrogen fixation of nitrogenase, which will be further explained below. | ||
==Ecology== | ==Ecology== | ||
''Halorhodospira halophila'' | ''Halorhodospira halophila's'' photoactive yellow protein (PYP) is a blue-light sensor that, upon detection of blue light, induces the organism to swim away. [12] It also functions to serve as an oxidizer of sulfide to sulfur, which then is stored outside of the cell where it is oxidized further into sulfate. Sulfur oxidization by phototrophic bacteria is also depend on ecological and psychiological factors, as speciazation of sulfur groups in different categories of sulfur-oxidizing bacteria is shown to be present. [14] | ||
As explained above, ''Halorhodospira halophila'' also has a role in hydrogen production through the nitrogen fixation process with nitrogenase. | |||
==Pathology== | ==Pathology== | ||
| Line 73: | Line 73: | ||
==Application to Biotechnology== | ==Application to Biotechnology== | ||
As will be explained below, current research has speculated that ''Halorhodospira halophila'' could play a strong role in nitrogen fixation through | As will be explained below, current research has speculated that ''Halorhodospira halophila'' could play a strong role in hydrogen generation as a byproduct of nitrogen fixation through nitrogenase. Research in this field can aide in the use of the organism in photobiological hydrogen generation to develop new fuel cells as a possible "next-generation fuel source". [9] | ||
'Halorhodospira halophila' also serves in the production of organic solutes such as glycine betaine, ectoine, and trehalose, which act to help balance osmotic pressure. The organism also helps with the oxidization of sulfide to sulfur, which after being deposited outside of the cell can then be oxidized into sulfate. [2] | |||
==Current Research== | ==Current Research== | ||
| Line 86: | Line 87: | ||
[http://www.jstage.jst.go.jp/article/jbb/101/3/263/_pdf Cloning and Characterization of nif Structural and Regulatory Genes in the Purple Sulfur Bacterium, Halorhodospira halophila] | [http://www.jstage.jst.go.jp/article/jbb/101/3/263/_pdf Cloning and Characterization of nif Structural and Regulatory Genes in the Purple Sulfur Bacterium, Halorhodospira halophila] | ||
===Photoactive Yellow Protein (PYP) used in research=== | |||
Much of the current research involving ''Halorhodospira halophila'' specifically focuses on the blue-light sensor PYP that is present. PYP belongs to the Xanthopsins, "a family of blue-light photoreceptors that contain 4-hydroxy-cinnamic acid as their photoactive chromophore". As a small protein, PYP serves as a good model system for researching how a chromophore and protein interact "to sense light and send a biological signal." [4] One such research aspect is the use of PYP from ''Halorhodospira halophila'' to analyze the photocycle process of PYP to understand its complex mechanism, each of 5 detailed intermediates, etc. [11] PYP in ''Halorhodospira halophila'' is important because the mechanism causes a signaled response to blue-light and induces movement of the organism. Current research in this field analyzes how each step of the signaling process works. [12] This PYP has become important because it has become the "structural prototype for the PAS domain, and plays a role in many signaling pathways. [13] | |||
Several aspects of research are currently being conducted on the function and attributes of PYP: | |||
[http://www.pnas.org/cgi/content/full/103/41/15050 Ultrafast infrared spectroscopy reveals a key step for successful entry into the photocycle for photoactive yellow protein] | |||
[http://www.biophysj.org/cgi/content/abstract/87/3/1858?ijkey=eb9a238116c9c974bb0f1f27eb55ee8137e7f612&keytype2=tf_ipsecsha Incoherent Manipulation of the Photoactive Yellow Protein Photocycle with Dispersed Pump-Dump-Probe Spectroscopy] | |||
[http://www.pnas.org/cgi/content/abstract/102/20/7145?ijkey=4fa750d180a08415544bf91d9f7775773a026713&keytype2=tf_ipsecsha Visualizing reaction pathways in photoactive yellow protein from nanoseconds to seconds] | |||
[http://pubs.acs.org/cgi-bin/abstract.cgi/bichaw/2003/42/i34/abs/bi034878p.html Initial steps of signal generation in photoactive yellow protein revealed with femtosecond mid-infrared spectroscopy] | |||
[http://www.rsc.org/delivery/_ArticleLinking/DisplayArticleForFree.cfm?doi=b315731h&JournalCode=PP Photoactive yellow protein, bacteriophytochrome, and sensory rhodopsin in purple phototrophic bacteria] | |||
==References== | ==References== | ||
| Line 108: | Line 124: | ||
10. [http://www.springerlink.com/content/7b3j189vdcl1aryl/ Imhoff, J.F. and Suling, Jorg. "''The phylogenetic relationship among Ectothiorhodospiraceae: a reevaluation of their taxonomy on the basis of 16S rDNA analyses''". ''Archives of Microbiology''. Springerlink. 19 February, 2004. June 2007.] | 10. [http://www.springerlink.com/content/7b3j189vdcl1aryl/ Imhoff, J.F. and Suling, Jorg. "''The phylogenetic relationship among Ectothiorhodospiraceae: a reevaluation of their taxonomy on the basis of 16S rDNA analyses''". ''Archives of Microbiology''. Springerlink. 19 February, 2004. June 2007.] | ||
11. [http://www.pnas.org/cgi/content/abstract/102/20/7145?ijkey=4fa750d180a08415544bf91d9f7775773a026713&keytype2=tf_ipsecsha Ihee, Hyotcherl et al. "''Visualizing reaction pathways in photoactive yellow protein from nanoseconds to seconds''". ''PNAS.com''. Proceedings of the National Academy of Sciences of the United States of America. 3 May 2005. June 2007.] | |||
12. [http://pubs.acs.org/cgi-bin/abstract.cgi/bichaw/2003/42/i34/abs/bi034878p.html Groot, M.L. et al. "''Initial steps of signal generation in photoactive yellow protein revealed with femtosecond mid-infrared spectroscopy''". ''Biochemistry''. American Chemical Society. 5 August, 2003. June 2007.] | |||
13. [http://www.rsc.org/publishing/journals/PP/article.asp?doi=b315731h Kyndt, John et al. "''Photoactive yellow protein, bacteriophytochrome, and sensory''". ''Photochemical and Photobiological Sciences''. RSC Publishing. 20 April 2004. June 2007.] | |||
rhodopsin in purple phototrophic bacteria] | |||
14. [http://mic.sgmjournals.org/cgi/reprint/148/1/267 Prange, Alexander et al. "''Quantitative speciation of sulfur in bacterial sulfur globules: X-ray absorption spectroscopy reveals at least three different species of sulfur''". ''Microbiology''. 2002. June 2007.] | |||
Edited by student Kent Lee of [mailto:ralarsen@ucsd.edu Rachel Larsen] and Kit Pogliano | Edited by student Kent Lee of [mailto:ralarsen@ucsd.edu Rachel Larsen] and Kit Pogliano | ||
Revision as of 17:39, 8 June 2007
A Microbial Biorealm page on the genus Halorhodospira halophila
Classification
Higher order taxa
Superkingdom: Bacteria;
Phylum: Proteobacteria;
Class: Gammaproteobacteria;
Order: Chromatiales;
Family: Ectothiorhodospiraceae;
Genus: Halorhodospira;
Species: Halorhodospira halophila
Species
Halorhodospira halophila
|
NCBI: Taxonomy |
Strains
Halorhodospira halophila DSM 244/S1
Halorhodospira halophila BN9624
Halorhodospira halophila BN9630
Description and significance
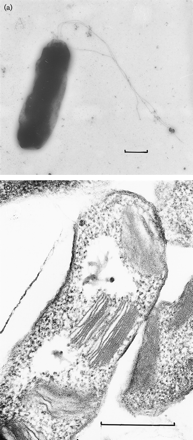
Halorhodospira halophila (formerly Ectothiorhodospira halophila) is an extremely halophilic purple bacterium that was formally a member of the Ectothiorhodospira genus until recently reclassified. Phylogenetically, Halorhodospira halohila is associated within the gamma subdivision of the phylum Proteobacteria and is known to be phototrophic and Gram-negative. It is considered to be “one of the most halophilic eubacteria known.” [2] Its halophilic nature allows for it to be present in conditions that often have been thought to be too harsh for bacteria, and it is found proliferating in saturated salts such as crystallizer ponds (a hypersaline environment where sodium chloride precipitates) that have a salinity of 25% or higher. This importantly shows that harsh environments once thought to be exclusive to archaea actually contain bacteria as well. [3]
This halophilic organism was first isolated from a salt lake mud [5] and has been determined to have several functions: production of organic solutes glycine betaine, ectoine, and trehalose to help balance osmotic pressure, as well as the oxidization of sulfide to sulfur (which later would be further oxidized into sulfate). [2] This has resulted in the use of Halorhodospira halophila for research on topics from photobiological ability to produce hydrogen gas to effects on sulfur.
One important aspect is the production of the bacterial photoreceptor PYP (photoactive yellow protein) within Halorhodospira halophila. PYP is a blue-light sensor found in Halorhodospira halophila that has great significance towards protein research and biotechnology. Photoactive proteins are generally accepted as "model systems for studying protein signal state formation." As a small protein, PYP provides for an 'attractive model system for exploring how a chromophore and protein interact to sense light and send a biological signal." [4]
Halorhodospira halophila has one major strain (DSM244/SL1) whose genome has been sequenced. But two other strains are recognized: BN9624 and BN9630. [1]
Genome structure
The gene sequence of Halorhodospira halophila S1 is the only listed strain of the species that has fully been sequenced. Genome sequencing of Halorhodospira halophila S1 was completed in January 2007 by the Department of Energy Joint Genome Institute. The genome is 2,678,452 nucleotides long (1,339,226 base pairs) and is made up of circular DNA. There are 2493 genes, 2407 which are protein coding, as well as 55 structural RNAs and 31 pseudo genes. The GC content of strain S1 is 67.9%. There is no current information on plasmids related to this species. [6]
While not all strains of Halorhodospira halophila have been sequenced, there is still some information on their similarity to related species. Halorhodospira neutriphila, displayed in the photo on the right, has a 94.6% similarity to strain DSM 244. Meanwhile, similarities with strains BN 9624 and BN 9630 are 91.4% and 90.7% respectively.
Cell structure and metabolism
Halorhodospira halophila is known as a "purple sulfur bacterium", whose structure consists of two membranes as well as the presence of flagella. [2] Its size is about ~4Mb. [7]
Halorhodospira halophila is capable of a large number of metabolic pathways such as glycolysis, the citrate cylce, amino acid metabolism, and more. A full list of these pathways and their individual maps can be found at the KEGG Pathway Database for Halorhodospira halophila. [8]
One important part of Halorhodospira halophila (and subject to recent research) is the presence of Photoactive Yellow Protein (PYP), a 14kDa cytoplasmic photoreceptor protein, which is an important protein for current research.
Halorhodospira halophila also has a potential role of hydrogen production through the nitrogen fixation of nitrogenase, which will be further explained below.
Ecology
Halorhodospira halophila's photoactive yellow protein (PYP) is a blue-light sensor that, upon detection of blue light, induces the organism to swim away. [12] It also functions to serve as an oxidizer of sulfide to sulfur, which then is stored outside of the cell where it is oxidized further into sulfate. Sulfur oxidization by phototrophic bacteria is also depend on ecological and psychiological factors, as speciazation of sulfur groups in different categories of sulfur-oxidizing bacteria is shown to be present. [14]
As explained above, Halorhodospira halophila also has a role in hydrogen production through the nitrogen fixation process with nitrogenase.
Pathology
At this time, Halorhodospira halophila causes no known diseases.
Application to Biotechnology
As will be explained below, current research has speculated that Halorhodospira halophila could play a strong role in hydrogen generation as a byproduct of nitrogen fixation through nitrogenase. Research in this field can aide in the use of the organism in photobiological hydrogen generation to develop new fuel cells as a possible "next-generation fuel source". [9]
'Halorhodospira halophila' also serves in the production of organic solutes such as glycine betaine, ectoine, and trehalose, which act to help balance osmotic pressure. The organism also helps with the oxidization of sulfide to sulfur, which after being deposited outside of the cell can then be oxidized into sulfate. [2]
Current Research
Designation of Halorhodospira as a separate genus
Recent studies of the sequences of the 16sRNA gene have been able to provide details and data into the Ectothiorhodospira genus and the different species within, helping to distinguish Halorhodospira and reassigning it as a separate genus category. Within the Ectothiorhodospira genus, three organisms with an extremely halophilic nature showed enough difference to warrant reclassification: Halorhodospira halophila, Halorhodospira halochloris, and Halorhodospira abdelmalekii.[10]
Photobiological hydrogen generation
Some of the recent research on Halorhodospira halophila includes focusing on it's photobiological ability to produce hydrogen gas and the potential applications of this. Through the nitrogen fixation process in photosynthetic bacteria involving nitrogenase, hydrogen generation typically results as a byproduct. However, most of the studies of photobiological hydrogen production has been with purple non-sulfur bacteria. Current research is looking into the hydrogen production capabilities specific to purple sulfur bacteria. Purple sulfur bacteria are preferable for hydrogen generation because they are "complete photoautotrophs and can be cultivated in minimal culture media." Additionally, Halorhodospira halophila's" capability to withstand high pH and salt environments is a benefit since seawater could be "used directly as a culture medium". Current research, as shown by the paper below, shows that Halorhodospira halophila is capable of generating a substantial amount of hydrogen, and thus is a viable candidate for helping in the production of renewable energy sources in the future. [9]
Photoactive Yellow Protein (PYP) used in research
Much of the current research involving Halorhodospira halophila specifically focuses on the blue-light sensor PYP that is present. PYP belongs to the Xanthopsins, "a family of blue-light photoreceptors that contain 4-hydroxy-cinnamic acid as their photoactive chromophore". As a small protein, PYP serves as a good model system for researching how a chromophore and protein interact "to sense light and send a biological signal." [4] One such research aspect is the use of PYP from Halorhodospira halophila to analyze the photocycle process of PYP to understand its complex mechanism, each of 5 detailed intermediates, etc. [11] PYP in Halorhodospira halophila is important because the mechanism causes a signaled response to blue-light and induces movement of the organism. Current research in this field analyzes how each step of the signaling process works. [12] This PYP has become important because it has become the "structural prototype for the PAS domain, and plays a role in many signaling pathways. [13]
Several aspects of research are currently being conducted on the function and attributes of PYP:
Visualizing reaction pathways in photoactive yellow protein from nanoseconds to seconds
References
9. [http://www.jstage.jst.go.jp/article/jbb/101/3/263/_pdf Hisayoshi Tsuihiji, Yoichi Yamazaki, Hironari Kamikubo, Yasushi Imamoto and Mikio Kataoka: “Cloning and Characterization of nif Structural and Regulatory Genes in the Purple Sulfur Bacterium, Halorhodospira halophila”. J. BIOSCI. BIOENG., Vol. 101, 263-270 (2006).
13. Kyndt, John et al. "Photoactive yellow protein, bacteriophytochrome, and sensory". Photochemical and Photobiological Sciences. RSC Publishing. 20 April 2004. June 2007. rhodopsin in purple phototrophic bacteria]
14. Prange, Alexander et al. "Quantitative speciation of sulfur in bacterial sulfur globules: X-ray absorption spectroscopy reveals at least three different species of sulfur". Microbiology. 2002. June 2007. Edited by student Kent Lee of Rachel Larsen and Kit Pogliano
