Dental Plaque Biofilms: Difference between revisions
No edit summary |
|||
| (92 intermediate revisions by one other user not shown) | |||
| Line 1: | Line 1: | ||
{{Curated}} | |||
By: Anna Frutiger | By: Anna Frutiger | ||
==Introduction== | ==Introduction== | ||
<br>Dental plaque can be defined as a diverse community of microorganisms | <br>[[Image:gingivitis.gif|200 x 150 px|left|thumb|Photo of someone exhibiting gingivitis [http://www.thejcdp.com/issue003/overman/01over.htm<4>]]] | ||
<br>Dental plaque is commonly known as the primary cause of dental caries and other oral infections. Rightly so, as despite the ever increasing knowledge in oral health care, the average adult in the United States has between 10 and 17 decayed, missing or filled permanent teeth [4]. Additionally, the majority of the population has at least minor gingivitis, an inflammation of the gums, with a much smaller percentage suffering from moderate to severe periodontitis [4]. Dental plaque, which exists not only on the tooth surface but also under the gums, can be defined as a diverse community of microorganisms in the form of a biofilm. The microorganisms bind tightly to one another, in addition to the solid tooth surface, by means of an extracellular matrix consisting of polymers of both host and microbial origin [1][2]. Only recently has dental plaque begun to be considered a biofilm, which has further contributed to a better understanding of common oral diseases and has helped to find the best ways to prevent and control these for the future [3]. | |||
As a biofilm, dental plaque exhibits an open architecture much like that of other biofilms. The open architechture, which consists of channels and voids, helps to achieve the flow of nutrients, waste products, metabolites, enzymes, and oxygen through the biofilm [4]. Because of this structure, a variety of microbial organisms can make up biofilms, including both aerobic and anaerobic bacteria. The microbial composition of dental biofilms includes over 700 species of bacteria and archaea, which all exist in a relatively stable environment called microbial homeostasis [2]. Dental plaque biofilms are responsible for many of the diseases common to the oral cavity including dental caries, periodontitis, gingivitis, and the less common peri-implantitis (similar to periodontitis, but with dental implants), however biofilms are present on healthy teeth as well [5]. | |||
<br>[[Image:Structuralbiofilm.gif|450 × 200 px|right|thumb|Structural representation of a dental plaque biofilm [http://www.thejcdp.com/issue003/overman/01over.htm<4>]]] | |||
The microbial make-up of the normal plaque biofilm, however, differs immensely from the biofilm formed in carious regions and in the sub-gingival regions associated with periodontal disease. Because bacteria in dental biofilms (and other biofilms) express an entirely different set of genes than free-floating bacteria, they are of particular interest to researchers and thus much is currently being done to explore the diverse nature of these microbial structures. The biofilm state brings about major changes in bacterial behavior such as interactions with other microbes and the host, in addition to responding to changing environmental conditions [5]. It was the incredible antibiotic resistance of plaque biofilms that originally sparked the notion that dental plaque was not formed by free-floating bacteria, as plaque removal had to be done via mechanical means, rather than just with antibiotics. Because of these behavioral changes, in addition to the microbial differences present between healthy and diseased biofilms, much of the research currently being done is in relation to plaque-related diseases and cell-cell communication within the dental biofilm and the oral cavity. This is of major concern to the medical community, because it is being realized that oral pathologies have a major impact not only on the health of the oral cavity, but also on the overall health of the human body. | |||
<br> | |||
==Formation of Dental Biofilms== | ==Formation of Dental Biofilms== | ||
<br>The formation of dental plaque biofilms includes a series of steps that | <br>The formation of dental plaque biofilms includes a series of steps that begins with the initial colonization of the pellicle and ends with the complex formation of a mature biofilm. Dental plaque biofilms exist on a variety of tooth surfaces including fissues, smooth surfaces and gingival crevices, however they are most likely to be seen in their mature state in the more stagnant sites, like fissures and crevices, as these places provide protection from the forces of removal, like a toothbrush [1][5]. Additionally, through the growth process of the plaque biofilm, the microbial composition changes from one that is primarily gram-positive and streptococcus-rich to a structure filled with gram-negative anaerobes in its more mature state [6]. <br> | ||
===Adsorption of Host and Bacterial Molecules to the Tooth Surface=== | ===Adsorption of Host and Bacterial Molecules to the Tooth Surface=== | ||
<br>The first step in plaque biofilm development is the adsorption of host and bacterial molecules to the tooth surface. Within minutes of tooth eruption or a cleaning, pellicle formation begins, which | <br>The first step in plaque biofilm development is the adsorption of host and bacterial molecules to the tooth surface. Within minutes of tooth eruption or a cleaning, pellicle formation begins, which can be defined as a thin coat of salivary proteins [3]. The pellicle acts like an adhesive by sticking to the tooth surface and encouraging a conditioning film of bacteria to attach to the pellicle [1]. This conditioning film directly influences the initial microbial colonization, and continues to adsorb bacteria to the tooth surface. | ||
Healthy tooth surfaces and gingivae tend to only be associated with this first phase of biofilm development. It consists of an initial few layers (1-20) of mostly gram-positive cocci bacteria, followed by some gram-positive rods and fillaments and a very small amount of gram-negative cocci. The gram-positive cocci species involved in this conditioning layer include <i>[[Streptococcus mutans]]</i>, <i>[[Streptococcus mitis]]</i>, <i>[[Streptococcus]] sanguis</i>, <i>[[Streptococcus]] oralis</i>, <i>[[Rothia dentocariosa]]</i>, and <i>[[Staphylococcus epidermidis]]</i>. The gram-negative rod and filament species | Healthy tooth surfaces and gingivae tend to only be associated with this first phase of biofilm development. It consists of an initial few layers (1-20) of mostly gram-positive cocci bacteria, followed by some gram-positive rods and fillaments and a very small amount of gram-negative cocci. The gram-positive cocci species involved in this conditioning layer include, but are not limited to, <i>[[Streptococcus mutans]]</i>, <i>[[Streptococcus mitis]]</i>, <i>[[Streptococcus]] sanguis</i>, <i>[[Streptococcus]] oralis</i>, <i>[[Rothia dentocariosa]]</i>, and <i>[[Staphylococcus epidermidis]]</i>. The gram-negative rod and filament species include <i>[[Actinomyces viscosus]]</i>, <i>[[Actinomyces israelis]]</i>. <i>[[Actinomyces gerencseriae]]</i> and the <i>[[Corynebacterium]]</i> species. <i>[[Veillonella parvula]]</i> and the <i> [[Neisseria]]</i> species make up some of the few gram-negative cocci, which are aerobes or facultative aerobes and are able to adhere to the non-exfoliating hard tooth surfaces [5]. This early composition of the biofilm is able to withstand many of the frequent mechanisms of the oral cavity that contribute to bacterial removal such as swallowing, nose blowing, chewing, and salivary fluid outflow. The early colonizers are also able to survive in the high oxygen concentrations present in the oral cavity, without having much protection from other bacteria [5]. Thus, this thin, initial biofilm is almost always present on the tooth surface as it forms immediately after cleaning. | ||
<br> | <br> | ||
| Line 22: | Line 30: | ||
<br>Following pellicle formation, there is passive transport of oral bacteria to the tooth surface, which involves a reversible adhesion process [2]. By using weak, long-range physicochemical interactions between the pellicle coated tooth surface and the microbial cell surface, an area of weak attraction is formed that encourages the reversible adhesion [1]. This reversible adhesion then leads to a much stronger, irreversible attachment, as short-range interactions between specific molecules on the bacterial cells and the complementary receptor proteins on the pellicle surface occur. Because many oral | <br>Following pellicle formation, there is passive transport of oral bacteria to the tooth surface, which involves a reversible adhesion process [2]. By using weak, long-range physicochemical interactions between the pellicle coated tooth surface and the microbial cell surface, an area of weak attraction is formed that encourages the microbes to reverse their previous adhesion to the pellicle and come off the tooth surface (hence the term "reversible adhesion") [1]. This reversible adhesion then leads to a much stronger, irreversible attachment, as short-range interactions between specific molecules on the bacterial cells and the complementary receptor proteins on the pellicle surface occur. Because many oral microbial species have multiple adhesion types on their cell surface, they can thus participate in a plethora of interactions with both other microbes and with the host surface molecules [1]. <br> | ||
===Co-Adhesion of later colonizers to already attached early colonizers=== | ===Co-Adhesion of later colonizers to already attached early colonizers=== | ||
[[Image:corncob.gif|250 × 100 px|right|thumb|SEM of the corn-cob morphological structure present on plaque biofilms. Photo courtesy of Charles Cobb. [http://www.thejcdp.com/issue003/overman/01over.htm<4>]]] | [[Image:corncob.gif|250 × 100 px|right|thumb|Figure 1: SEM of the corn-cob morphological structure present on plaque biofilms. Photo courtesy of Charles Cobb. [http://www.thejcdp.com/issue003/overman/01over.htm<4>]]] | ||
<br>The co-adhesion of the later colonizers to the already present biofilm continues to involve many specific interactions between bacterial receptors and adhesions. These interactions build up the biofilm to create a more diverse environment, which includes the development of unusual morphological structures like corn-cobs and rosettes [1] (Figure 1). | |||
The many interactions between these diverse bacterial species begin to create a number of synergistic and antagonistic biochemical interactions. For example, bacterial residing in food chains may help to contribute metabolically with other bacteria if they are physically | The many interactions between these diverse bacterial species begin to create a number of synergistic and antagonistic biochemical interactions. For example, bacterial residing in food chains may help to contribute metabolically with other bacteria if they are located physically close to one another. Similarly, when obligate anaerobes and aerobes are involved in co-adhesion, these interactions can ensure the anaerobic bacteria’s survival in the oxygen-rich oral cavity [1]. | ||
<br> | |||
===Multiplication of the Attached Microorganisms=== | ===Multiplication of the Attached Microorganisms=== | ||
<br> | |||
Eventually, the bacterial cells continue to divide until a three-dimensional mixed-culture biofilm forms that is spacially and functionally organized. Polymer production causes the development of the extracellular matrix, which consists of soluble and insoluble glucans, fructans, and heteropolymers [1]. This matrix is one of the key structural aspects of the plaque biofilm, much like that of other biofilms. Biofilms such as this are very thick, consisting of 100-300 cell layers. The bacterial stratification is arranged according to metabolism and aerotolerance, with the number of gram-negative cocci, rods and filaments increasing as more anaerobic bacteria appear [5]. As the biofilm thickens and becomes more mature, these anaerobic bacteria can live deeper within the biofilm, to further protect them from the oxygen-rich environment within the oral cavity. | |||
<br> | |||
==Inter-species Communication== | ==Inter-species Communication== | ||
<br> | <br>The discovery that communication between cells in biofilm communities occurs has been key in understanding how dental plaque acts as a single unit. Communication can occur in a variety of ways, including gene expression, cell-cell signaling (ex. quorum sensing), and antibiotic resistance, among others. Specific bacteria within the biofilm community are able to act with other species to both help and impair the host, in addition to providing a positive cooperation between the different species of the biofilm [5]. Further, the patterns observed of microbial colonization and coaggregation appear to be primarily uni-directional, thus indicating that many of the bacterial species in dental biofilms require an environment that has been previously habituated by other microbiota in order to properly colonize [5]. These specific cell-cell interactions have proven to be very important topics of current research involving dental plaque biofilms. | ||
== | [[Image:Cluster.gif|500 x 400 px|left|thumb|Figure 2: Microbial complexes arranged into clusters, as proposed by Socransky and Haffajee. [http://www3.interscience.wiley.com/cgi-bin/fulltext/118702639/HTMLSTART<7>]]] | ||
<br> | |||
Sigmund Socransky and Anne Haffajee have been able to explain much of the unique colonization patterns and positive cooperation observed in plaque biofilm. The method that has helped explain this is one involving “clustered” groups of bacteria, where each cluster is created based on similarities and differences in nutritional and atmospheric environments [5][7](Figure 2). Each of the clusters is said to influence the other clusters, in addition to being related to a specific periodontal state, thus showing that these microbes are very closely associated with one another [5]. Socransky and Haffajee found that within different periodontal states, either all or none of the species belonging to a particular cluster were found. Very few individual species or pairs of species were found, which further helped show that biofilms were a community of microbes. Certain clusters were also found to interact particularly well with other clusters. For example, microbial species found in the red cluster were rarely observed without the presence of the orange cluster species [5]. Because of these associations found by Socransky and Haffajee and their cluster analysis, it provided further knowledge to the idea that the microbial communities in plaque biofilms were involved in a great deal of inter-species communication. | |||
[[Image:Figure1.jpg|300 x 250 px|right|thumb|Figure 3: (a) In the presence of <i>S. cristatus</i> CC5A <i>P. gingivalis</i> was inhibited from colonizing on the plaque biofilm. (b) In the presence of <i>S. gordonii</i> G9B, however, colonization of <i>P. gingivalis</i> occurred, thus indicating that <i>S. gordonii</i> was not involved in a cell signaling mechanism similar to <i>S. cristatus</i>. Red = Streptococcal cells, Green = <i>P. gingivalis</i>, and yellow = colonized bacteria. [http://jb.asm.org/cgi/reprint/182/24/7067<6>]]] | |||
One such example of this involves the gram-negative, anaerobic bacteria <i>[[Porphyromonas gingivalis]]</i>. <i>P. gingivalis</i> is a predominant pathogen found in severe cases of adult periodontitis that attaches to the tooth surface after the initial colonization of the gram-positive <i>[[Streptococcus]]</i> species. The colonization of <i>P. gingivalis</i> is key in the transformation of the bacterial biofilm from a commensal plaque to the pathogenic form observed in periodontitis [6]. Its colonization is also dependent on its attachment to oral surfaces, which is accomplished via a fimbriae-mediated adhesion. The <i>fimA</i> gene encodes for this fimbriae subunit protein, which can be regulated via environmental conditions [6]. Interestingly, <i>P. gingivalis</i> <i>fimA</i> gene expression was found to be severely downregulated by the unrelated, gram-positive <i>[[Streptococcus cristatus]]</i> CC5A, which can be found in the initial colonization of dental biofilms. By interfering with the gene that encodes for the fimbriae-subunit protein, <i>S. cristatus</i> is able to prevent <i>P. gingivalis</i> fimbriae attachment to the biofilm, thus resulting in an easier removal of <i>P. gingivalis</i> from the biofilm by salivary flow (Figure 3). This study was performed by introducing <i>P. gingivalis</i> to plates of both <i>S. cristatus</i> (Figure 3a) and <i>S. gordonii</i>, which is another key component of early plaque (Figure 3b). From the figure, it is clear that on the <i>S. cristatus</i> plate, <i>P. gingivalis</i> failed to bind to or accumulate to the <i>S. cristatus</i> cells. On the <i>S. gordonii</i> cell plate, however, <i>P. gingivalis</i>bound to and accumulated in biofilm microcolonies. From these results, it was clear that <i>S. cristatus</i> had an antagonistic signaling interaction with <i>P. gingivalis</i>, thus preventing <i>P. gingivalis</i> colonization. After further testing, it was discovered that the <i>S. cristatus</i> signaling activity was specific to the <i>fimA</i> gene, and is also associated with a 59-kDa surface protein. The size of the surface protein, however, suggests that it is inherently distinct from the signaling peptides that are secreted via other gram-positive bacteria [6]. | |||
Through these findings, it was clear that some signaling mechanisms between <i>P. gingivalis</i> and <i>S. cristatus</i> were occurring, which were preventing the colonization of <i>P. gingivalis</i> on the plaque biofilm. Of course, it is likely that other microbial species also contribute to the upregulation of this gene, and thus may negatively affect the downregulation of <i>fimA </i> caused by <i>S. cristatus</i>. It is also known that <i>Fusobacterium nucleatum</i> is involved with some type of cell-sigaling interaction with <i>P. gingivalis</i>, but in a different way. Rather than affecting fimbriae attachment, <i>F. nucleatum</i> is instead required for the <i>P. gingivalis</i> aggregation with the facultative aerobes already present on the biofilm [5]. This directly shows that inter-species communication between the different microbial species on dental plaque biofilm is required for proper biofilm maturation and is thus directly involved in oral disease propagation as well. This research should give us a better understanding of the regulation of <i>P. gingivalis</i> via cell-cell interactions within the dental plaque biofilm. | |||
[[Image:Quorumsensing.gif|250 x 200 px|left|thumb|Figure 4: SEM of <i>S. mutans</i> wild-type (A and C) and <i>comC</i> mutant (B and D) biofilm formation. Magnification (A and B): 480x; Magnification (C and D): 4,800x. [http://jb.asm.org/cgi/content/full/184/10/2699?view=long&pmid=11976299<8>]]] | |||
Yet another interesting aspect of cell-cell signaling on dental plaque biofilm deals with quorum sensing in <i>[[Streptococcus mutans]]</i>. Quorum sensing, in the general sense, is a mechanism that bacteria employ to control gene expression in response to the population size [8]. While it had been previously known that quorum sensing in gram-positive bacteria regulates genetic competence development, it was unclear what the involvement of this was during signaling in biofilm initiation and formation. In particular, it was unclear whether biofilm formation of <i>S. mutans</i>, which has particular importance in dental plaque biofilms, was involved in any type of coordinated activity through cell-cell communication [8]. By mutating various aspects of the quorum-sensing and signal-transduction systems, it was discovered that the competence-stimulating peptide (CSP) was necessary for proper biofilm formation of <i>S. mutans</i>. In particular, the <i>comC</i> mutant, which was unable to produce or secrete CSP, resulted in biofilms formed with an altered architecture compared to those of the wildtype (Figure 4). From looking at this figure, it is clear that the <i>comC</i> mutant biofilms had an obviously disrupted structure (Fig. 4B and D) than those of the wildtype (Fig. 4A and C), as the mutant photographs display gaps of disorganization within the biofilm. Additionally, the study showed that <i>comD, comE</i> and <i>comX</i> mutants also resulted in abnormal biofilm formation, as they were formed with a reduced biomass. Because four different CSP mutants caused an improper biofilm formation by <i>S. mutans</i>, it is likely that CSP affects multiple signal-transduction pathways. It was also shown that this CSP-mediated quorum-sensing causes the transformation frequency of <i>S. mutans</i> be 10- to 600- fold greater in biofilms than in planktonic cells [1][8]. This shows that the quorum-sensing pathway in <i>S. mutans </i> needed for genetic competence is key in the proper formation of dental plaque biofilms [8]. | |||
<br> | |||
==Microbial Biofilm Composition of Oral Disease States== | |||
<br>Many studies have been performed that attempt to determine which bacterial species are directly involved in oral pathology. Because many of the plaque-mediated oral diseases occur at regions already containing an extremely diverse microflora, it is difficult to exactly specify which of these species are pathogenic. Additionally, the bacterial traits associated with cariogenicity (acid production, acid tolerance, intracellular and extracellular polysaccharide production) point to more than a single bacterial species [2]. We do know, however, that many of the desirable bacterial species involved in healthy plaque biofilms include <i>Streptococcus sanguis</i>, <i> S. gordonii</i>, <i>S. oralis</i>, and the <i>Actinomyces</i> species, in addition to other related bacteria with a low acid tolerance [9]. Therefore, it seems that healthy dental biofilm microflora consists of species with limited tolerance for acid, as we do know that the bacteria involved in the formation of dental caries are those with a very high acid tolerance. <br> | |||
===Caries=== | |||
[[Image:cariestable.gif|550 x 300 px|right|thumb| Table 1: Acid tolerance diversity characteristics of four dental plaque bacterial species. [http://www.springerlink.com/content/u453787t4h143252/fulltext.pdf <9>]]] | |||
<br>Despite the lack of exact knowledge on every pathogen involved in caries production, the factors responsible for microbial homeostasis within the biofilm are known and recognized [2]. The initial change in environment is due to an increased amount of fermentable carbohydrates in the diet of the host. The anaerobic, acid-producing bacteria present in the plaque biofilm thus produce an increased amount of acid due to fermentaion, consequently lowering the pH of the biofilm [2][9]. When the pH drops, there is an increase in these acid-tolerant bacteria, as they are the only ones that can survive and perform glycolysis in such acidic environments. Some of the more common bacterial species responsible for this include <i>Streptococcus mutans</i>, <i>S. sorbrinus</i>, and <i>Lactobacillus casei</i> which can perform glycolysis at a pH level as low as 3.0 [9]. Table 1 demonstrates the acid tolerance characteristics of a select number of bacteria involved in the dental plaque biofilm and shows the vast differences that exist between normal oral microflora species. At this acidic pH, the hightly acid-tolerant bacterial biofilm is capable of demineralizing the tooth enamel, with greater degrees of acidity causing faster rates of demineralization. Of course, this acidification is originally caused by sugar ingestion, meaning if sugar intake stops, the pH value of the biofilm will rise again and remineralization of the enamel can occur. Caries will result, however, if the acidification-demineralization phase is more damaging and more frequent than the alkalinization-remineralization phase can manage to fix the damage [9]. The demineralization of tooth enamel can also occur solely from the presence of highly acidic substances in the oral cavity. This is why people who drink excessive amounts of soda pop (with a highly acidic pH of 2.3-5) have a much higher prevalence of caries. In short, when sugar is ingested and acid is produced as a metabolic biproduct, bacteria that can survive in these acidic environments will thrive. These are the key pathogens to caries production. It is important to recognize, however, that some of the known bacterial species involved in caries production, like <i>S. mutans</i>, are also present in healthy plaque biofilm, in addition to also being absent from some sites of caries production. Thus, we know that there is not one particular bacterial species responsible for caries production, but a collection of several, exhibiting similar characteristics [2]. <br> | |||
===Periodontitis and Peri-implantitis=== | |||
<br>Some level of periodontal disease affects a majority of the adult population of the United States. Because of this, it is of great importance to the medical and dental community and can thus be considered a public health problem [5]. Periodontitis, if not treated early, can lead to alveolar bone and tooth loss. It is defined by deep pockets formed between the tooth surface and the gum, with this deep pocket being easily colonized by microbes due to the small dentinal tubules and enamel fissues that lead directly into the gums from the open space of the mouth [9]. This area is incredibly difficult to reach via typical oral health care mechanisms (tooth brush, floss, etc.), which often leads to the diseased states of gingivitis or periodontitis, that have varying levels of severity. | |||
[[Image:bargraph.gif|550 x 400 px|left|thumb| Figure 5: Bar graphs of the prevelence of spirochetes and <i>P. gingivalis</i> in oral sites and patients with differing periodontal health. [http://www3.interscience.wiley.com/cgi-bin/fulltext/118702639/HTMLSTART<7>]]] | |||
Much of the microflora existing in these deep periodontal pockets are gram-negative anaerobes, with a very diverse population of spirochetes. In the early stages of periodontal disease, known as gingivitis, the initial microbial colonization of the plaque biofilm seems to involve members of the yellow, green and purple “clusters” (Figure 2) [7]. Secondary colonization occurs with members of the orange and red clusters, and these become more dominant. The increased levels of the red and orange cluster bacteria lead to proliferation by members of all the original and secondary colonizing species. At a certain point, the organisms must disperse to other locations within the oral cavity to ensure survival. A study found that spirochetes and <i>P. gingivalis</i> were more prevalent in diseased sites of diseased patients than in healthy sites of diseased patients. It was also found that the organisms were found more frequently in healthy sites of diseased patients than in healthy sites of healthy patients, which is evidence for the dispersal mechanism previously mentioned (Figure 5)[7]. The bar graphs presented in Figure 5 show the frequency of spirochetes and <i>P. gingivalis</i> present in various dental decay sites in patients ranging from healthy to having advanced periodontitis. The graph clearly supports the information presented above, as bacterial levels were remarkably higher in diseased patients. | |||
[[Image:DNADNA1.gif|350 x 350 px|right|thumb| Figure 6: Checkerboard DNA-DNA hybridization analysis that was used to detect the presence of 40 microbial species in 28 subgingival plaque biofilm samples. [http://www3.interscience.wiley.com/cgi-bin/fulltext/118702639/HTMLSTART<7>]]] | |||
As periodontal disease gets more severe, checkerboard DNA-DNA hybridization experiments have been performed to give a better idea of the species involved in periodontitis (Figure 6). This molecular biology experiment was used to detect the presence of various bacteiral species by using known DNA probes on the horizontal lanes, and plaque samples from a number of patients in the vertical lanes. By looking at the blot, it is clear which bacterial species were present in the plaque in these periodontal patients, as the DNA probes bound to their corresponding DNA sequences of bacteria present in the plaque. Further analysis has been performed from this, to show that the most prevalent bacterial species involved in periodontitis is <i>Actinomyces naeslundii</i>. These tests were performed on 40 species of which there were known molecular probes. Unfortunately, there is no real way to find every single bacterial species involved in the plaque biofilm of periodontitis patients, as a molecular probe is needed for checkerboard DNA-DNA hybridization, and we do not have known probes for all of these species. [7]. | |||
Peri-implantitis is very similar to periodontitis, however, it does differ in some aspects. Because dental implants are not surrounded by periodontal ligaments, they have differing biomechanics and defensive cell-recruitment [5]. Peri-implantitis refers to the destruction of the supporting peri-implant tissue due to a microbial infection. These infections tend to occur around places where residual teeth or failing implants can act as reservoirs for bacteria and form biofilm colonies. Interestingly, the bacterial species involved in peri-implantitis are very similar to those that play a key role in periodontitis[5]. The two diseases differ in some key ways, but they do have many similarities and research in both can help lead to better treatment and prevention. | |||
<br> | |||
==Conclusion== | ==Conclusion== | ||
<br> | |||
<br>Dental plaque biofilms are a diverse, functioning microbial community that are found on every organism on earth that has teeth. Because of the wide diversity of organisms involved in the development and proper function of plaque biofilm, it is difficult to know everything there is to know about these fascinating microbial communities. These biofilms employ a great deal of inter-cell communication to not only keep themselves alive, but also to protect the host. Their existence, while involved in many pathogenic oral diseases, is of much benefit to the host at the early stages of development, as it provides the teeth with a layer of protection that cannot be matched. In time, we will continue to discover more about the biochemical and developmental functions involved in dental plaque biofilms, which can help us to not only learn about microbiology, but also to improve oral health, which is of great importance to our overall well-being. <br> | |||
==References== | ==References== | ||
| Line 58: | Line 117: | ||
[http://jb.asm.org/cgi/content/full/182/24/7067?view=long&pmid=11092870 6] Hua, X., Cook, G. S., Costerton, J. W., Bruce, G., Rose, T. M., Lamont, R. J. "Intergeneric Communication in Dental Plaque Biofilms." <i>Journal of Bacteriology</i>. 2000. Volume 182. p. 7067-7069. | [http://jb.asm.org/cgi/content/full/182/24/7067?view=long&pmid=11092870 6] Hua, X., Cook, G. S., Costerton, J. W., Bruce, G., Rose, T. M., Lamont, R. J. "Intergeneric Communication in Dental Plaque Biofilms." <i>Journal of Bacteriology</i>. 2000. Volume 182. p. 7067-7069. | ||
[http://www3.interscience.wiley.com/cgi-bin/fulltext/118702639/HTMLSTART 7] Socransky, S. S., Haffajee, A. D., "Periodontal Microbial Ecology." <i>Periodontology 2000</i>. Volume 38. p. 135-187. | |||
[http://jb.asm.org/cgi/content/full/184/10/2699?view=long&pmid=11976299 8] Li, Y., Tang, N., Aspiras, M. B., Lau, P. C. Y., Lee, J. H., Ellen, R. P., Cvitkovitch, D. G. "A Quorum-Sensing Signaling System Essential for Genetic Competence in <i>Streptococcus mutans</i> Is Involved in Biofilm Formation. <i> Journal of Bacteriology</i>. 2002. Volume 184. p. 2699-2708. | |||
[http://www.springerlink.com/content/u453787t4h143252/fulltext.pdf 9] Marquis, M. E., "Oxygen metabolism, oxidative stress adn acid-base physiology of dental plaque biofilms." <i>Journal of Industrial Microbiology</i>. Volume 15. p. 198-207. | |||
Edited by [mailto:frutigera@kenyon.edu Anna Frutiger], student of [mailto:slonczewski@kenyon.edu Joan Slonczewski] for [http://biology.kenyon.edu/courses/biol238/biol238syl09.html BIOL 238 Microbiology], 2009, [http://www.kenyon.edu/index.xml Kenyon College]. | Edited by [mailto:frutigera@kenyon.edu Anna Frutiger], student of [mailto:slonczewski@kenyon.edu Joan Slonczewski] for [http://biology.kenyon.edu/courses/biol238/biol238syl09.html BIOL 238 Microbiology], 2009, [http://www.kenyon.edu/index.xml Kenyon College]. | ||
Latest revision as of 20:13, 10 August 2010
By: Anna Frutiger
Introduction
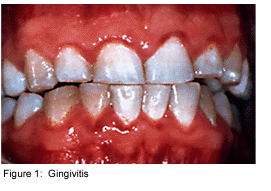
Dental plaque is commonly known as the primary cause of dental caries and other oral infections. Rightly so, as despite the ever increasing knowledge in oral health care, the average adult in the United States has between 10 and 17 decayed, missing or filled permanent teeth [4]. Additionally, the majority of the population has at least minor gingivitis, an inflammation of the gums, with a much smaller percentage suffering from moderate to severe periodontitis [4]. Dental plaque, which exists not only on the tooth surface but also under the gums, can be defined as a diverse community of microorganisms in the form of a biofilm. The microorganisms bind tightly to one another, in addition to the solid tooth surface, by means of an extracellular matrix consisting of polymers of both host and microbial origin [1][2]. Only recently has dental plaque begun to be considered a biofilm, which has further contributed to a better understanding of common oral diseases and has helped to find the best ways to prevent and control these for the future [3].
As a biofilm, dental plaque exhibits an open architecture much like that of other biofilms. The open architechture, which consists of channels and voids, helps to achieve the flow of nutrients, waste products, metabolites, enzymes, and oxygen through the biofilm [4]. Because of this structure, a variety of microbial organisms can make up biofilms, including both aerobic and anaerobic bacteria. The microbial composition of dental biofilms includes over 700 species of bacteria and archaea, which all exist in a relatively stable environment called microbial homeostasis [2]. Dental plaque biofilms are responsible for many of the diseases common to the oral cavity including dental caries, periodontitis, gingivitis, and the less common peri-implantitis (similar to periodontitis, but with dental implants), however biofilms are present on healthy teeth as well [5].
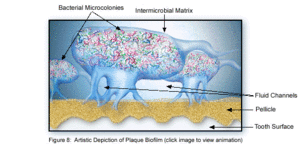
The microbial make-up of the normal plaque biofilm, however, differs immensely from the biofilm formed in carious regions and in the sub-gingival regions associated with periodontal disease. Because bacteria in dental biofilms (and other biofilms) express an entirely different set of genes than free-floating bacteria, they are of particular interest to researchers and thus much is currently being done to explore the diverse nature of these microbial structures. The biofilm state brings about major changes in bacterial behavior such as interactions with other microbes and the host, in addition to responding to changing environmental conditions [5]. It was the incredible antibiotic resistance of plaque biofilms that originally sparked the notion that dental plaque was not formed by free-floating bacteria, as plaque removal had to be done via mechanical means, rather than just with antibiotics. Because of these behavioral changes, in addition to the microbial differences present between healthy and diseased biofilms, much of the research currently being done is in relation to plaque-related diseases and cell-cell communication within the dental biofilm and the oral cavity. This is of major concern to the medical community, because it is being realized that oral pathologies have a major impact not only on the health of the oral cavity, but also on the overall health of the human body.
Formation of Dental Biofilms
The formation of dental plaque biofilms includes a series of steps that begins with the initial colonization of the pellicle and ends with the complex formation of a mature biofilm. Dental plaque biofilms exist on a variety of tooth surfaces including fissues, smooth surfaces and gingival crevices, however they are most likely to be seen in their mature state in the more stagnant sites, like fissures and crevices, as these places provide protection from the forces of removal, like a toothbrush [1][5]. Additionally, through the growth process of the plaque biofilm, the microbial composition changes from one that is primarily gram-positive and streptococcus-rich to a structure filled with gram-negative anaerobes in its more mature state [6].
Adsorption of Host and Bacterial Molecules to the Tooth Surface
The first step in plaque biofilm development is the adsorption of host and bacterial molecules to the tooth surface. Within minutes of tooth eruption or a cleaning, pellicle formation begins, which can be defined as a thin coat of salivary proteins [3]. The pellicle acts like an adhesive by sticking to the tooth surface and encouraging a conditioning film of bacteria to attach to the pellicle [1]. This conditioning film directly influences the initial microbial colonization, and continues to adsorb bacteria to the tooth surface.
Healthy tooth surfaces and gingivae tend to only be associated with this first phase of biofilm development. It consists of an initial few layers (1-20) of mostly gram-positive cocci bacteria, followed by some gram-positive rods and fillaments and a very small amount of gram-negative cocci. The gram-positive cocci species involved in this conditioning layer include, but are not limited to, Streptococcus mutans, Streptococcus mitis, Streptococcus sanguis, Streptococcus oralis, Rothia dentocariosa, and Staphylococcus epidermidis. The gram-negative rod and filament species include Actinomyces viscosus, Actinomyces israelis. Actinomyces gerencseriae and the Corynebacterium species. Veillonella parvula and the Neisseria species make up some of the few gram-negative cocci, which are aerobes or facultative aerobes and are able to adhere to the non-exfoliating hard tooth surfaces [5]. This early composition of the biofilm is able to withstand many of the frequent mechanisms of the oral cavity that contribute to bacterial removal such as swallowing, nose blowing, chewing, and salivary fluid outflow. The early colonizers are also able to survive in the high oxygen concentrations present in the oral cavity, without having much protection from other bacteria [5]. Thus, this thin, initial biofilm is almost always present on the tooth surface as it forms immediately after cleaning.
Passive Transport of Oral Bacteria to the Tooth Surface
Following pellicle formation, there is passive transport of oral bacteria to the tooth surface, which involves a reversible adhesion process [2]. By using weak, long-range physicochemical interactions between the pellicle coated tooth surface and the microbial cell surface, an area of weak attraction is formed that encourages the microbes to reverse their previous adhesion to the pellicle and come off the tooth surface (hence the term "reversible adhesion") [1]. This reversible adhesion then leads to a much stronger, irreversible attachment, as short-range interactions between specific molecules on the bacterial cells and the complementary receptor proteins on the pellicle surface occur. Because many oral microbial species have multiple adhesion types on their cell surface, they can thus participate in a plethora of interactions with both other microbes and with the host surface molecules [1].
Co-Adhesion of later colonizers to already attached early colonizers
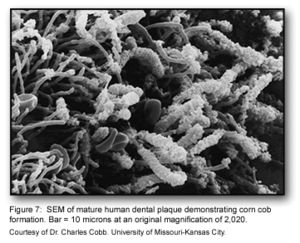
The co-adhesion of the later colonizers to the already present biofilm continues to involve many specific interactions between bacterial receptors and adhesions. These interactions build up the biofilm to create a more diverse environment, which includes the development of unusual morphological structures like corn-cobs and rosettes [1] (Figure 1).
The many interactions between these diverse bacterial species begin to create a number of synergistic and antagonistic biochemical interactions. For example, bacterial residing in food chains may help to contribute metabolically with other bacteria if they are located physically close to one another. Similarly, when obligate anaerobes and aerobes are involved in co-adhesion, these interactions can ensure the anaerobic bacteria’s survival in the oxygen-rich oral cavity [1].
Multiplication of the Attached Microorganisms
Eventually, the bacterial cells continue to divide until a three-dimensional mixed-culture biofilm forms that is spacially and functionally organized. Polymer production causes the development of the extracellular matrix, which consists of soluble and insoluble glucans, fructans, and heteropolymers [1]. This matrix is one of the key structural aspects of the plaque biofilm, much like that of other biofilms. Biofilms such as this are very thick, consisting of 100-300 cell layers. The bacterial stratification is arranged according to metabolism and aerotolerance, with the number of gram-negative cocci, rods and filaments increasing as more anaerobic bacteria appear [5]. As the biofilm thickens and becomes more mature, these anaerobic bacteria can live deeper within the biofilm, to further protect them from the oxygen-rich environment within the oral cavity.
Inter-species Communication
The discovery that communication between cells in biofilm communities occurs has been key in understanding how dental plaque acts as a single unit. Communication can occur in a variety of ways, including gene expression, cell-cell signaling (ex. quorum sensing), and antibiotic resistance, among others. Specific bacteria within the biofilm community are able to act with other species to both help and impair the host, in addition to providing a positive cooperation between the different species of the biofilm [5]. Further, the patterns observed of microbial colonization and coaggregation appear to be primarily uni-directional, thus indicating that many of the bacterial species in dental biofilms require an environment that has been previously habituated by other microbiota in order to properly colonize [5]. These specific cell-cell interactions have proven to be very important topics of current research involving dental plaque biofilms.
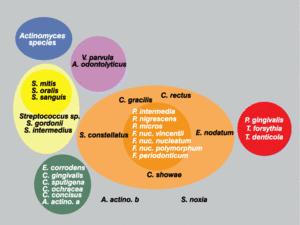
Sigmund Socransky and Anne Haffajee have been able to explain much of the unique colonization patterns and positive cooperation observed in plaque biofilm. The method that has helped explain this is one involving “clustered” groups of bacteria, where each cluster is created based on similarities and differences in nutritional and atmospheric environments [5][7](Figure 2). Each of the clusters is said to influence the other clusters, in addition to being related to a specific periodontal state, thus showing that these microbes are very closely associated with one another [5]. Socransky and Haffajee found that within different periodontal states, either all or none of the species belonging to a particular cluster were found. Very few individual species or pairs of species were found, which further helped show that biofilms were a community of microbes. Certain clusters were also found to interact particularly well with other clusters. For example, microbial species found in the red cluster were rarely observed without the presence of the orange cluster species [5]. Because of these associations found by Socransky and Haffajee and their cluster analysis, it provided further knowledge to the idea that the microbial communities in plaque biofilms were involved in a great deal of inter-species communication.
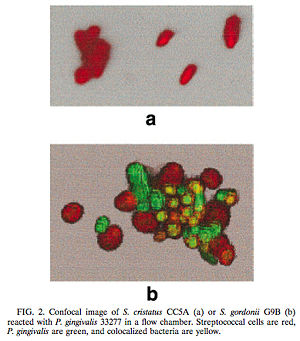
One such example of this involves the gram-negative, anaerobic bacteria Porphyromonas gingivalis. P. gingivalis is a predominant pathogen found in severe cases of adult periodontitis that attaches to the tooth surface after the initial colonization of the gram-positive Streptococcus species. The colonization of P. gingivalis is key in the transformation of the bacterial biofilm from a commensal plaque to the pathogenic form observed in periodontitis [6]. Its colonization is also dependent on its attachment to oral surfaces, which is accomplished via a fimbriae-mediated adhesion. The fimA gene encodes for this fimbriae subunit protein, which can be regulated via environmental conditions [6]. Interestingly, P. gingivalis fimA gene expression was found to be severely downregulated by the unrelated, gram-positive Streptococcus cristatus CC5A, which can be found in the initial colonization of dental biofilms. By interfering with the gene that encodes for the fimbriae-subunit protein, S. cristatus is able to prevent P. gingivalis fimbriae attachment to the biofilm, thus resulting in an easier removal of P. gingivalis from the biofilm by salivary flow (Figure 3). This study was performed by introducing P. gingivalis to plates of both S. cristatus (Figure 3a) and S. gordonii, which is another key component of early plaque (Figure 3b). From the figure, it is clear that on the S. cristatus plate, P. gingivalis failed to bind to or accumulate to the S. cristatus cells. On the S. gordonii cell plate, however, P. gingivalisbound to and accumulated in biofilm microcolonies. From these results, it was clear that S. cristatus had an antagonistic signaling interaction with P. gingivalis, thus preventing P. gingivalis colonization. After further testing, it was discovered that the S. cristatus signaling activity was specific to the fimA gene, and is also associated with a 59-kDa surface protein. The size of the surface protein, however, suggests that it is inherently distinct from the signaling peptides that are secreted via other gram-positive bacteria [6].
Through these findings, it was clear that some signaling mechanisms between P. gingivalis and S. cristatus were occurring, which were preventing the colonization of P. gingivalis on the plaque biofilm. Of course, it is likely that other microbial species also contribute to the upregulation of this gene, and thus may negatively affect the downregulation of fimA caused by S. cristatus. It is also known that Fusobacterium nucleatum is involved with some type of cell-sigaling interaction with P. gingivalis, but in a different way. Rather than affecting fimbriae attachment, F. nucleatum is instead required for the P. gingivalis aggregation with the facultative aerobes already present on the biofilm [5]. This directly shows that inter-species communication between the different microbial species on dental plaque biofilm is required for proper biofilm maturation and is thus directly involved in oral disease propagation as well. This research should give us a better understanding of the regulation of P. gingivalis via cell-cell interactions within the dental plaque biofilm.
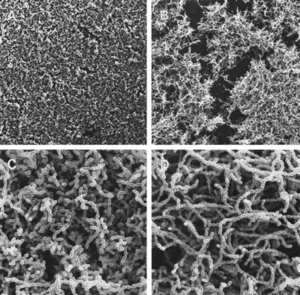
Yet another interesting aspect of cell-cell signaling on dental plaque biofilm deals with quorum sensing in Streptococcus mutans. Quorum sensing, in the general sense, is a mechanism that bacteria employ to control gene expression in response to the population size [8]. While it had been previously known that quorum sensing in gram-positive bacteria regulates genetic competence development, it was unclear what the involvement of this was during signaling in biofilm initiation and formation. In particular, it was unclear whether biofilm formation of S. mutans, which has particular importance in dental plaque biofilms, was involved in any type of coordinated activity through cell-cell communication [8]. By mutating various aspects of the quorum-sensing and signal-transduction systems, it was discovered that the competence-stimulating peptide (CSP) was necessary for proper biofilm formation of S. mutans. In particular, the comC mutant, which was unable to produce or secrete CSP, resulted in biofilms formed with an altered architecture compared to those of the wildtype (Figure 4). From looking at this figure, it is clear that the comC mutant biofilms had an obviously disrupted structure (Fig. 4B and D) than those of the wildtype (Fig. 4A and C), as the mutant photographs display gaps of disorganization within the biofilm. Additionally, the study showed that comD, comE and comX mutants also resulted in abnormal biofilm formation, as they were formed with a reduced biomass. Because four different CSP mutants caused an improper biofilm formation by S. mutans, it is likely that CSP affects multiple signal-transduction pathways. It was also shown that this CSP-mediated quorum-sensing causes the transformation frequency of S. mutans be 10- to 600- fold greater in biofilms than in planktonic cells [1][8]. This shows that the quorum-sensing pathway in S. mutans needed for genetic competence is key in the proper formation of dental plaque biofilms [8].
Microbial Biofilm Composition of Oral Disease States
Many studies have been performed that attempt to determine which bacterial species are directly involved in oral pathology. Because many of the plaque-mediated oral diseases occur at regions already containing an extremely diverse microflora, it is difficult to exactly specify which of these species are pathogenic. Additionally, the bacterial traits associated with cariogenicity (acid production, acid tolerance, intracellular and extracellular polysaccharide production) point to more than a single bacterial species [2]. We do know, however, that many of the desirable bacterial species involved in healthy plaque biofilms include Streptococcus sanguis, S. gordonii, S. oralis, and the Actinomyces species, in addition to other related bacteria with a low acid tolerance [9]. Therefore, it seems that healthy dental biofilm microflora consists of species with limited tolerance for acid, as we do know that the bacteria involved in the formation of dental caries are those with a very high acid tolerance.
Caries
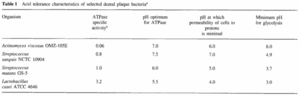
Despite the lack of exact knowledge on every pathogen involved in caries production, the factors responsible for microbial homeostasis within the biofilm are known and recognized [2]. The initial change in environment is due to an increased amount of fermentable carbohydrates in the diet of the host. The anaerobic, acid-producing bacteria present in the plaque biofilm thus produce an increased amount of acid due to fermentaion, consequently lowering the pH of the biofilm [2][9]. When the pH drops, there is an increase in these acid-tolerant bacteria, as they are the only ones that can survive and perform glycolysis in such acidic environments. Some of the more common bacterial species responsible for this include Streptococcus mutans, S. sorbrinus, and Lactobacillus casei which can perform glycolysis at a pH level as low as 3.0 [9]. Table 1 demonstrates the acid tolerance characteristics of a select number of bacteria involved in the dental plaque biofilm and shows the vast differences that exist between normal oral microflora species. At this acidic pH, the hightly acid-tolerant bacterial biofilm is capable of demineralizing the tooth enamel, with greater degrees of acidity causing faster rates of demineralization. Of course, this acidification is originally caused by sugar ingestion, meaning if sugar intake stops, the pH value of the biofilm will rise again and remineralization of the enamel can occur. Caries will result, however, if the acidification-demineralization phase is more damaging and more frequent than the alkalinization-remineralization phase can manage to fix the damage [9]. The demineralization of tooth enamel can also occur solely from the presence of highly acidic substances in the oral cavity. This is why people who drink excessive amounts of soda pop (with a highly acidic pH of 2.3-5) have a much higher prevalence of caries. In short, when sugar is ingested and acid is produced as a metabolic biproduct, bacteria that can survive in these acidic environments will thrive. These are the key pathogens to caries production. It is important to recognize, however, that some of the known bacterial species involved in caries production, like S. mutans, are also present in healthy plaque biofilm, in addition to also being absent from some sites of caries production. Thus, we know that there is not one particular bacterial species responsible for caries production, but a collection of several, exhibiting similar characteristics [2].
Periodontitis and Peri-implantitis
Some level of periodontal disease affects a majority of the adult population of the United States. Because of this, it is of great importance to the medical and dental community and can thus be considered a public health problem [5]. Periodontitis, if not treated early, can lead to alveolar bone and tooth loss. It is defined by deep pockets formed between the tooth surface and the gum, with this deep pocket being easily colonized by microbes due to the small dentinal tubules and enamel fissues that lead directly into the gums from the open space of the mouth [9]. This area is incredibly difficult to reach via typical oral health care mechanisms (tooth brush, floss, etc.), which often leads to the diseased states of gingivitis or periodontitis, that have varying levels of severity.
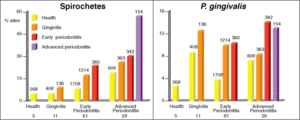
Much of the microflora existing in these deep periodontal pockets are gram-negative anaerobes, with a very diverse population of spirochetes. In the early stages of periodontal disease, known as gingivitis, the initial microbial colonization of the plaque biofilm seems to involve members of the yellow, green and purple “clusters” (Figure 2) [7]. Secondary colonization occurs with members of the orange and red clusters, and these become more dominant. The increased levels of the red and orange cluster bacteria lead to proliferation by members of all the original and secondary colonizing species. At a certain point, the organisms must disperse to other locations within the oral cavity to ensure survival. A study found that spirochetes and P. gingivalis were more prevalent in diseased sites of diseased patients than in healthy sites of diseased patients. It was also found that the organisms were found more frequently in healthy sites of diseased patients than in healthy sites of healthy patients, which is evidence for the dispersal mechanism previously mentioned (Figure 5)[7]. The bar graphs presented in Figure 5 show the frequency of spirochetes and P. gingivalis present in various dental decay sites in patients ranging from healthy to having advanced periodontitis. The graph clearly supports the information presented above, as bacterial levels were remarkably higher in diseased patients.

As periodontal disease gets more severe, checkerboard DNA-DNA hybridization experiments have been performed to give a better idea of the species involved in periodontitis (Figure 6). This molecular biology experiment was used to detect the presence of various bacteiral species by using known DNA probes on the horizontal lanes, and plaque samples from a number of patients in the vertical lanes. By looking at the blot, it is clear which bacterial species were present in the plaque in these periodontal patients, as the DNA probes bound to their corresponding DNA sequences of bacteria present in the plaque. Further analysis has been performed from this, to show that the most prevalent bacterial species involved in periodontitis is Actinomyces naeslundii. These tests were performed on 40 species of which there were known molecular probes. Unfortunately, there is no real way to find every single bacterial species involved in the plaque biofilm of periodontitis patients, as a molecular probe is needed for checkerboard DNA-DNA hybridization, and we do not have known probes for all of these species. [7].
Peri-implantitis is very similar to periodontitis, however, it does differ in some aspects. Because dental implants are not surrounded by periodontal ligaments, they have differing biomechanics and defensive cell-recruitment [5]. Peri-implantitis refers to the destruction of the supporting peri-implant tissue due to a microbial infection. These infections tend to occur around places where residual teeth or failing implants can act as reservoirs for bacteria and form biofilm colonies. Interestingly, the bacterial species involved in peri-implantitis are very similar to those that play a key role in periodontitis[5]. The two diseases differ in some key ways, but they do have many similarities and research in both can help lead to better treatment and prevention.
Conclusion
Dental plaque biofilms are a diverse, functioning microbial community that are found on every organism on earth that has teeth. Because of the wide diversity of organisms involved in the development and proper function of plaque biofilm, it is difficult to know everything there is to know about these fascinating microbial communities. These biofilms employ a great deal of inter-cell communication to not only keep themselves alive, but also to protect the host. Their existence, while involved in many pathogenic oral diseases, is of much benefit to the host at the early stages of development, as it provides the teeth with a layer of protection that cannot be matched. In time, we will continue to discover more about the biochemical and developmental functions involved in dental plaque biofilms, which can help us to not only learn about microbiology, but also to improve oral health, which is of great importance to our overall well-being.
References
1 Marsh, P.D. "Dental Plaque as a Microbial Biofilm." Caries Research. 2004. Volume 38. p.204-211.
2 Marsh, P.D. "Dental plaque as a biofilm and a microbial community - implications for health and disease." BMC Oral Health. 2006. 6(Suppl 1): S14.
3 Nield-Gehrig, J. S. "Dental Plaque Biofilms." Dental Plaque Biofilms.
4 Overman, P. R. "Biofilm: A New View of Plaque." The Journal of Contemporary Dental Practice. 2000. Volume 1. No. 3. P. 018.
5 Sbordone, L., Bortolaia, C. "Oral microbial biofilms and plaque-related diseases: microbial communities and their role in the shift from oral health to disease." Clin Oral Invest. 2003. Volume 7. P. 181-188.
6 Hua, X., Cook, G. S., Costerton, J. W., Bruce, G., Rose, T. M., Lamont, R. J. "Intergeneric Communication in Dental Plaque Biofilms." Journal of Bacteriology. 2000. Volume 182. p. 7067-7069.
7 Socransky, S. S., Haffajee, A. D., "Periodontal Microbial Ecology." Periodontology 2000. Volume 38. p. 135-187.
8 Li, Y., Tang, N., Aspiras, M. B., Lau, P. C. Y., Lee, J. H., Ellen, R. P., Cvitkovitch, D. G. "A Quorum-Sensing Signaling System Essential for Genetic Competence in Streptococcus mutans Is Involved in Biofilm Formation. Journal of Bacteriology. 2002. Volume 184. p. 2699-2708.
9 Marquis, M. E., "Oxygen metabolism, oxidative stress adn acid-base physiology of dental plaque biofilms." Journal of Industrial Microbiology. Volume 15. p. 198-207.
Edited by Anna Frutiger, student of Joan Slonczewski for BIOL 238 Microbiology, 2009, Kenyon College.
