Bacillus selenatarsenatis: Difference between revisions
Thomas355263 (talk | contribs) |
Thomas355263 (talk | contribs) |
||
| (23 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
{{Uncurated}} | |||
== Classification == | == Classification == | ||
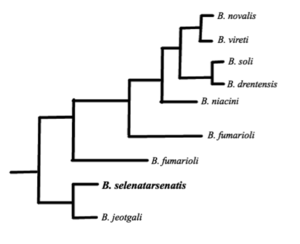
===Higher | [[Image:Phy.png |thumb|300px|right|Figure 1: Phylogenic tree of Bacillus selenatarsenatis based on the 16S rRNA sequence. This diagram is adapted from Yamamura, S. ''et al'' [[#References|[2]]] ]] | ||
Bacteria; Firmicutes; Bacilli; Bacillales; Bacillaceae; Bacillus | |||
===Higher Order Taxa=== | |||
Bacteria; Firmicutes; Bacilli; Bacillales; Bacillaceae; ''Bacillus'' | |||
=== Species === | === Species === | ||
| Line 13: | Line 17: | ||
''Bacillus selenatarsenatis'' | ''Bacillus selenatarsenatis'' | ||
The genome is not yet fully sequenced, but its genomic DNA has a GC content of 42.8%. [[#References|[2]]] Partial analysis that has been conducted on its [http://en.wikipedia.org/wiki/16S_ribosomal_RNA 16S rRNA] suggests that ''Bacillus jeotgali'' is its closest phylogenic relative. [[#References|[2]]] However, ''Bacillus jeotgali'' lacks the ability for dissimilatory reduction of selenate or arsenate. [[#References|[2]]] | |||
== Description == | == Description == | ||
Bacillus selenatarsenatis is a Gram-positive, oxidase-negative, catalase-positive, motile, spore-forming, rod-shaped, and facultatively anaerobic bacterium capable of respiring with selenate, arsenate, and nitrate as the terminal electron acceptor. [1] It was first isolated from a selenium-contaminated sediment obtained from an effluent drain of a glass-manufacturing plant in Japan. [2] It forms uniform red colonies, which indicates the formation of elemental selenium. [1] A few studies were conducted in attempt to utilize its selenate-reducing and arsenate-reducing properties for the removal of selenate and arsenate from the contaminated field via bioremediation. | ''Bacillus selenatarsenatis'' is a Gram-positive, oxidase-negative, catalase-positive, motile, spore-forming, rod-shaped, and facultatively anaerobic bacterium capable of respiring with selenate, arsenate, and nitrate as the terminal electron acceptor. [[#References|[1]]] It was first isolated from a selenium-contaminated sediment obtained from an effluent drain of a glass-manufacturing plant in Japan. [[#References|[2]]] It forms uniform red colonies, which indicates the formation of elemental selenium. [[#References|[1]]] A few studies were conducted in attempt to utilize its selenate-reducing and arsenate-reducing properties for the removal of selenate and arsenate from the contaminated field via bioremediation. | ||
== | == Metabolism == | ||
''B. selenatarsenatis'' can grow aerobically by [http://en.wikipedia.org/wiki/Oxidative_phosphorylation oxidative respiration], coupled with terminal electron acceptors like arsenate, selenate, or nitrate. Under anaerobic condition, ''B. selenatarsenatis'' can [http://en.wikipedia.org/wiki/Fermentation_%28biochemistry%29 ferment] various carbohydrates such as glucose, lactate, and pyruvate. [[#References|[2]]] ''B. selenatarsenatis'' is able to reduce selenate into selenite, then into elemental selenium; arsenate into arsenite; nitrate into nitrite, then into ammonia. [[#References|[1]]] | |||
Interestingly, the concentration of selenite (>2mM) has an inhibitory effect on the reduction of selenite to elemental selenium. [[#References|[1]]] | |||
Also, the presence of nitrate will have an inhibitory effect on the dissimilatory reduction of selenate and arsenate. [[#References|[1]]][[#References|[9]]] | |||
''B. selenatarsenatis'' has a unique [http://en.wikipedia.org/wiki/Operon operon] (srdBCA) that encodes a membrane-bound selenate reductase complex . [[#References|[5]]] This complex is consisted of three subunits: SrdA, SrdB, and SrdC. SrdC is a highly hydrophobic protein with nine transmembrane regions, which suggests that SrdC is an anchor protein that keeps the complex to the membrane. [[#References|[5]]] SrdB has an oxidoreductase activity that transfers electrons from quinone to SrdA. SrdA is also an oxidoreductase, which reduces selenate into selenite. [[#References|[5]]] Both SrdB and SrdA have four [4Fe-4S] clusters that are responsible for the electron transfers. [[#References|[5]]] Overall, this SrdBCA selenate reductase complex couples quinone oxidation with selenate reduction. | |||
Peroxiredoxin is also found in ''B. selenatarsenatis'', which suggests its role in reducing the toxicity of the selenium oxyanions by degrading the reactive oxygen species. [[#References|[10]]] | |||
== Ecology and Impact == | |||
[[Image:ArPoison.png |thumb|300px|right|Figure 2: Skin necrosis caused by arsenic poisoning. | |||
This image is from [http://manbir-online.com/diseases/arsenic.htm Manbir Online] ]] | |||
''B. selenatarsenatis'' is a mesophile with its optimal growth temperature between 25~40 degrees Celsius, and a pH between 7.5~9.0. [[#References|[1]]] | |||
Arsenate is a toxic inorganic compound that is analogous to phosphate and inhibits oxidative phosphorylation in many organisms. [3] It has a strong [http://en.wikipedia.org/wiki/Adsorption adsorption] to many common minerals found in sediments, which limits its solubility in water. ''B. selenatarsenatis'' is an arsenate-reducing organism that reduces arsenate into arsenite. Although both compounds are very toxic, the latter is more biologically available due to its relatively lower adsorption in comparison to arsenate. Arsenite can be more readily solubilized in water. [[#References|[4]]] It hinders the function of many proteins because of its strong affinity to sulfhydryl groups. [[#References|[3]]] Consequently, water sources will be contaminated by the toxic arsenite. Chronic intake of the contaminated water may result in arsenic poisoning. Arsenite leaching would be a concern for areas that are colonized by any arsenate-reducing organisms. | |||
It's natural niche is unknown, but it was first isolated in sediment outside of a glass manufacturing plant in Japan. | |||
== Bioremediation == | |||
[[Image:Spray.png|thumb|300px|right|Figure 3: Arsenate-based pesticide was being sprayed in a cotton field in 1927. | |||
Image attributed to [http://www.flickr.com/photos/usdagov/7087320559/in/set-72157627857689389/ U.S. Department of Agriculture]]] | |||
Before insecticide such as [http://en.wikipedia.org/wiki/Ddt DDT] was developed, lead arsenate was widely used as pesticide for tree fruit orchards. [[#References|[11]]] Arsenate-contaminated soil is problematic especially for agricultural land because plants will take up arsenate and accumulate it in the crops. These contaminated fields are difficult to treat because arsenate is not very soluble in water due to its strong adsorption with the minerals in the soil. [[#References|[4]]] Remediation using arsenate-reducing bacteria can convert arsenate into the more water-soluble arsenite, making extraction of arsenic easier. | |||
Selenate contamination in water can lead to serious consequences due to the high toxicity of selenate to organisms. Yet, there is no cost-effective chemical approach to remove selenate from water. [[#References|[6]]][[#References|[8]]] To solve this issue, a few researches were done on constructing a bioreactor containing ''B. selenatarsenatis'' to degrade selenate into selenite, which can be removed with an [http://en.wikipedia.org/wiki/Ion_exchanger ion exchanger] or further degraded by a few selenite degraders. [[#References|[6]]][[#References|[8]]] This is to prevent the inhibitory effect of selenite reduction of Bacillus selenatarsenatis from the selenite buildup. The researches showed promising results. | |||
== References == | |||
[http://www.sciencedirect.com/science/article/pii/S0922338X97811300 1.] Fujita, M., Ike, M., Nishimoto, S., Takahashi, K. & Kashiwa, M. (1997). Isolation and characterization of a novel selenate-reducing bacterium, Bacillus sp. SF-1. J Ferment Bioeng 83, 517–522. | |||
[http://http://ijsb.sgmjournals.org/content/57/5/1060.short 2.] Yamamura, S.; Yamashita, M.; Fujimoto, N.; Kuroda, M.; Kashiwa, M.; Sei, K.; Fujita, M.; Ike, M. Bacillus selenatarsenatis sp. nov., a selenate- and arsenate-reducing bacterium isolated from the effluent drain of a glass-manufacturing plant. Int. J. Syst. Evol. Microbiol 2007, 57, 1060–1064. | |||
[http://www.sciencemag.org/content/300/5621/939.short 3.] Oremland, R. S. & Stolz, J. F. (2003). The ecology of arsenic. Science 300, 939–944. | |||
[http://pubs.acs.org/doi/abs/10.1021/es703146f 4.] Yamamura, S., Watanabe, M., Kanzaki, M., Soda, S., & Ike, M. (2008). Removal of arsenic from contaminated soils by microbial reduction of arsenate and quinone. Environmental science & technology, 42(16), 6154-6159. | |||
[http://jb.asm.org/content/193/9/2141.short 5.] Kuroda, M., Yamashita, M., Miwa, E., Imao, K., Fujimoto, N., Ono, H., Kouta, N., Kazunari, S., & Ike, M. (2011). Molecular Cloning and Characterization of the srdBCA Operon, Encoding the Respiratory Selenate Reductase Complex, from the Selenate-Reducing Bacterium Bacillus selenatarsenatis SF-1. Journal of bacteriology, 193(9), 2141-2148. | |||
[http://www.sciencedirect.com/science/article/pii/S0011916411005571 6.] Soda, S., Kashiwa, M., Kagami, T., Kuroda, M., Yamashita, M., & Ike, M. (2011). Laboratory-scale bioreactors for soluble selenium removal from selenium refinery wastewater using anaerobic sludge. Desalination, 279(1), 433-438. | |||
[http://www.sciencedirect.com/science/article/pii/S0168165610009612 7.] Sakaguchi, T., Shimizu, R., & Mochida, Y. (2010). Isolation and characterization of marine selenium-oxyanion respiring bacteria for selenium recovery in saline environments. Journal of Biotechnology, 150, 223. | |||
[http://onlinelibrary.wiley.com/doi/10.1002/bit.10425/abstract 8.] Fujita, M., Ike, M., Kashiwa, M., Hashimoto, R., & Soda, S. (2002). Laboratory‐scale continuous reactor for soluble selenium removal using selenate‐reducing bacterium, Bacillus sp. SF‐1. Biotechnology and bioengineering, 80(7), 755-761. | |||
[http://http://ijsb.sgmjournals.org/content/57/5/1060.short 9.] Stolz, J. F., Basu, P., Santini, J. M., & Oremland, R. S. (2006). Arsenic and Selenium in Microbial Metabolism*. Annu. Rev. Microbiol., 60, 107-130. | |||
[http://aem.asm.org/content/77/13/4676.short 10.] Lenz, M., Kolvenbach, B., Gygax, B., Moes, S., & Corvini, P. F. (2011). Shedding light on selenium biomineralization: proteins associated with bionanominerals. Applied and environmental microbiology, 77(13), 4676-4680. | |||
[http://link.springer.com/article/10.1007%2FBF00483038?LI=true#page-1 11.] Peryea, F. J., & Creger, T. L. (1994). Vertical distribution of lead and arsenic in soils contaminated with lead arsenate pesticide residues. Water, Air, & Soil Pollution, 78(3), 297-306. | |||
11. Peryea, F. J., & Creger, T. L. (1994). Vertical distribution of lead and arsenic in soils contaminated with lead arsenate pesticide residues. Water, Air, & Soil Pollution, 78(3), 297-306. | |||
Latest revision as of 07:16, 14 December 2012
Classification
Higher Order Taxa
Bacteria; Firmicutes; Bacilli; Bacillales; Bacillaceae; Bacillus
Species
|
NCBI: Taxonomy |
Bacillus selenatarsenatis
The genome is not yet fully sequenced, but its genomic DNA has a GC content of 42.8%. [2] Partial analysis that has been conducted on its 16S rRNA suggests that Bacillus jeotgali is its closest phylogenic relative. [2] However, Bacillus jeotgali lacks the ability for dissimilatory reduction of selenate or arsenate. [2]
Description
Bacillus selenatarsenatis is a Gram-positive, oxidase-negative, catalase-positive, motile, spore-forming, rod-shaped, and facultatively anaerobic bacterium capable of respiring with selenate, arsenate, and nitrate as the terminal electron acceptor. [1] It was first isolated from a selenium-contaminated sediment obtained from an effluent drain of a glass-manufacturing plant in Japan. [2] It forms uniform red colonies, which indicates the formation of elemental selenium. [1] A few studies were conducted in attempt to utilize its selenate-reducing and arsenate-reducing properties for the removal of selenate and arsenate from the contaminated field via bioremediation.
Metabolism
B. selenatarsenatis can grow aerobically by oxidative respiration, coupled with terminal electron acceptors like arsenate, selenate, or nitrate. Under anaerobic condition, B. selenatarsenatis can ferment various carbohydrates such as glucose, lactate, and pyruvate. [2] B. selenatarsenatis is able to reduce selenate into selenite, then into elemental selenium; arsenate into arsenite; nitrate into nitrite, then into ammonia. [1]
Interestingly, the concentration of selenite (>2mM) has an inhibitory effect on the reduction of selenite to elemental selenium. [1] Also, the presence of nitrate will have an inhibitory effect on the dissimilatory reduction of selenate and arsenate. [1][9]
B. selenatarsenatis has a unique operon (srdBCA) that encodes a membrane-bound selenate reductase complex . [5] This complex is consisted of three subunits: SrdA, SrdB, and SrdC. SrdC is a highly hydrophobic protein with nine transmembrane regions, which suggests that SrdC is an anchor protein that keeps the complex to the membrane. [5] SrdB has an oxidoreductase activity that transfers electrons from quinone to SrdA. SrdA is also an oxidoreductase, which reduces selenate into selenite. [5] Both SrdB and SrdA have four [4Fe-4S] clusters that are responsible for the electron transfers. [5] Overall, this SrdBCA selenate reductase complex couples quinone oxidation with selenate reduction.
Peroxiredoxin is also found in B. selenatarsenatis, which suggests its role in reducing the toxicity of the selenium oxyanions by degrading the reactive oxygen species. [10]
Ecology and Impact
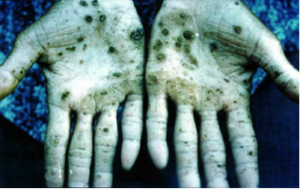
B. selenatarsenatis is a mesophile with its optimal growth temperature between 25~40 degrees Celsius, and a pH between 7.5~9.0. [1]
Arsenate is a toxic inorganic compound that is analogous to phosphate and inhibits oxidative phosphorylation in many organisms. [3] It has a strong adsorption to many common minerals found in sediments, which limits its solubility in water. B. selenatarsenatis is an arsenate-reducing organism that reduces arsenate into arsenite. Although both compounds are very toxic, the latter is more biologically available due to its relatively lower adsorption in comparison to arsenate. Arsenite can be more readily solubilized in water. [4] It hinders the function of many proteins because of its strong affinity to sulfhydryl groups. [3] Consequently, water sources will be contaminated by the toxic arsenite. Chronic intake of the contaminated water may result in arsenic poisoning. Arsenite leaching would be a concern for areas that are colonized by any arsenate-reducing organisms.
It's natural niche is unknown, but it was first isolated in sediment outside of a glass manufacturing plant in Japan.
Bioremediation

Before insecticide such as DDT was developed, lead arsenate was widely used as pesticide for tree fruit orchards. [11] Arsenate-contaminated soil is problematic especially for agricultural land because plants will take up arsenate and accumulate it in the crops. These contaminated fields are difficult to treat because arsenate is not very soluble in water due to its strong adsorption with the minerals in the soil. [4] Remediation using arsenate-reducing bacteria can convert arsenate into the more water-soluble arsenite, making extraction of arsenic easier.
Selenate contamination in water can lead to serious consequences due to the high toxicity of selenate to organisms. Yet, there is no cost-effective chemical approach to remove selenate from water. [6][8] To solve this issue, a few researches were done on constructing a bioreactor containing B. selenatarsenatis to degrade selenate into selenite, which can be removed with an ion exchanger or further degraded by a few selenite degraders. [6][8] This is to prevent the inhibitory effect of selenite reduction of Bacillus selenatarsenatis from the selenite buildup. The researches showed promising results.
References
1. Fujita, M., Ike, M., Nishimoto, S., Takahashi, K. & Kashiwa, M. (1997). Isolation and characterization of a novel selenate-reducing bacterium, Bacillus sp. SF-1. J Ferment Bioeng 83, 517–522.
2. Yamamura, S.; Yamashita, M.; Fujimoto, N.; Kuroda, M.; Kashiwa, M.; Sei, K.; Fujita, M.; Ike, M. Bacillus selenatarsenatis sp. nov., a selenate- and arsenate-reducing bacterium isolated from the effluent drain of a glass-manufacturing plant. Int. J. Syst. Evol. Microbiol 2007, 57, 1060–1064.
3. Oremland, R. S. & Stolz, J. F. (2003). The ecology of arsenic. Science 300, 939–944.
4. Yamamura, S., Watanabe, M., Kanzaki, M., Soda, S., & Ike, M. (2008). Removal of arsenic from contaminated soils by microbial reduction of arsenate and quinone. Environmental science & technology, 42(16), 6154-6159.
5. Kuroda, M., Yamashita, M., Miwa, E., Imao, K., Fujimoto, N., Ono, H., Kouta, N., Kazunari, S., & Ike, M. (2011). Molecular Cloning and Characterization of the srdBCA Operon, Encoding the Respiratory Selenate Reductase Complex, from the Selenate-Reducing Bacterium Bacillus selenatarsenatis SF-1. Journal of bacteriology, 193(9), 2141-2148.
6. Soda, S., Kashiwa, M., Kagami, T., Kuroda, M., Yamashita, M., & Ike, M. (2011). Laboratory-scale bioreactors for soluble selenium removal from selenium refinery wastewater using anaerobic sludge. Desalination, 279(1), 433-438.
7. Sakaguchi, T., Shimizu, R., & Mochida, Y. (2010). Isolation and characterization of marine selenium-oxyanion respiring bacteria for selenium recovery in saline environments. Journal of Biotechnology, 150, 223.
8. Fujita, M., Ike, M., Kashiwa, M., Hashimoto, R., & Soda, S. (2002). Laboratory‐scale continuous reactor for soluble selenium removal using selenate‐reducing bacterium, Bacillus sp. SF‐1. Biotechnology and bioengineering, 80(7), 755-761.
9. Stolz, J. F., Basu, P., Santini, J. M., & Oremland, R. S. (2006). Arsenic and Selenium in Microbial Metabolism*. Annu. Rev. Microbiol., 60, 107-130.
10. Lenz, M., Kolvenbach, B., Gygax, B., Moes, S., & Corvini, P. F. (2011). Shedding light on selenium biomineralization: proteins associated with bionanominerals. Applied and environmental microbiology, 77(13), 4676-4680.
11. Peryea, F. J., & Creger, T. L. (1994). Vertical distribution of lead and arsenic in soils contaminated with lead arsenate pesticide residues. Water, Air, & Soil Pollution, 78(3), 297-306.
