Odoribacter: Difference between revisions
No edit summary |
|||
| Line 10: | Line 10: | ||
::Species: ''Odoribacter denticanis'', ''Odoribacter laneus'', ''Odoribacter splanchnicus'' | ::Species: ''Odoribacter denticanis'', ''Odoribacter laneus'', ''Odoribacter splanchnicus'' | ||
=2. Description and significance= | =2. Description and significance= | ||
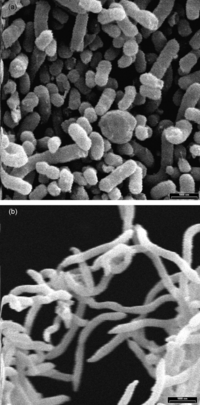
[[File:Odoribacter_Electron_Micrograph.gif|200px|thumb|right|Figure 1: Electron micrograph image of species belonging to the Odoribacter (bottom) [[#References|[9]]] ]] | |||
<blockquote> | <blockquote> | ||
The genus ''Odoribacter'' derives its name from its rod shape and foul odor it produces in the mouth of dogs [[#References |[1]]]. Bacteria within this genus are atypical opportunistic pathogens, anaerobic, Gram-negative, non-spore-forming, non-motile, catalase- and oxidase-negative [[#References |[1]]],[[#References|[2]]]. The ''Odoribacter'' was originally isolated from a ventral abscess in humans and classified as ''Bacteroides splanchnicus'' in 1971 [[#References |[1]]]. A new isolate that caused oral disease in companion animals was later found and classified as a new genus ''Odoribacter'' in 2008 [[#References |[1]]]. ''B. splanchnicus'' was then reclassified as ''O. splanchnicus'' [[#References |[1]]]. There are now three species within the genus: ''O. denticanis'', ''O. laneus'', and ''O. splanchnicus'' [[#References |[3]]]. Models for how each of these species operate ''in vivo''; however, have yet to be elucidated. Out of the three species, ''O. splanchnicus'' has been the most studied. | The genus ''Odoribacter'' derives its name from its rod shape and foul odor it produces in the mouth of dogs [[#References |[1]]]. Bacteria within this genus are atypical opportunistic pathogens, anaerobic, Gram-negative, non-spore-forming, non-motile, catalase- and oxidase-negative [[#References |[1]]],[[#References|[2]]]. The ''Odoribacter'' was originally isolated from a ventral abscess in humans and classified as ''Bacteroides splanchnicus'' in 1971 [[#References |[1]]]. A new isolate that caused oral disease in companion animals was later found and classified as a new genus ''Odoribacter'' in 2008 [[#References |[1]]]. ''B. splanchnicus'' was then reclassified as ''O. splanchnicus'' [[#References |[1]]]. There are now three species within the genus: ''O. denticanis'', ''O. laneus'', and ''O. splanchnicus'' [[#References |[3]]]. Models for how each of these species operate ''in vivo''; however, have yet to be elucidated. Out of the three species, ''O. splanchnicus'' has been the most studied. | ||
| Line 16: | Line 18: | ||
Members of the ''Odoribacter'' sp. can cause health problems such as abdominal abscess in humans and periodontal problems in domestic animals [[#References |[1]]],[[#References |[3]]]. More often, however, their presence is shown to be beneficial for the prevention of several diseases such as hypertension [[#References |[1]]],[[#References |[3]]]. As a member of the human intestinal microbiome, ''Odoribacter'' sp. can also modulate the host’s blood glucose concentration and blood pressure [[#References |[4]]],[[#References |[5]]].Thus, they are classified as atypical pathogens. | Members of the ''Odoribacter'' sp. can cause health problems such as abdominal abscess in humans and periodontal problems in domestic animals [[#References |[1]]],[[#References |[3]]]. More often, however, their presence is shown to be beneficial for the prevention of several diseases such as hypertension [[#References |[1]]],[[#References |[3]]]. As a member of the human intestinal microbiome, ''Odoribacter'' sp. can also modulate the host’s blood glucose concentration and blood pressure [[#References |[4]]],[[#References |[5]]].Thus, they are classified as atypical pathogens. | ||
</blockquote> | </blockquote> | ||
=3. Genome structure= | =3. Genome structure= | ||
<blockquote> | <blockquote> | ||
Revision as of 23:57, 12 December 2016
1. Classification
- a. Higher Order Taxa
- Domain: Bacteria
- Phylum: Bacteroidetes
- Class: Bacteroidia
- Order: Bacteroidales
- Family: Porphyromonoadaceae
- b. Species
- Species: Odoribacter denticanis, Odoribacter laneus, Odoribacter splanchnicus
2. Description and significance
The genus Odoribacter derives its name from its rod shape and foul odor it produces in the mouth of dogs [1]. Bacteria within this genus are atypical opportunistic pathogens, anaerobic, Gram-negative, non-spore-forming, non-motile, catalase- and oxidase-negative [1],[2]. The Odoribacter was originally isolated from a ventral abscess in humans and classified as Bacteroides splanchnicus in 1971 [1]. A new isolate that caused oral disease in companion animals was later found and classified as a new genus Odoribacter in 2008 [1]. B. splanchnicus was then reclassified as O. splanchnicus [1]. There are now three species within the genus: O. denticanis, O. laneus, and O. splanchnicus [3]. Models for how each of these species operate in vivo; however, have yet to be elucidated. Out of the three species, O. splanchnicus has been the most studied.
Members of the Odoribacter sp. can cause health problems such as abdominal abscess in humans and periodontal problems in domestic animals [1],[3]. More often, however, their presence is shown to be beneficial for the prevention of several diseases such as hypertension [1],[3]. As a member of the human intestinal microbiome, Odoribacter sp. can also modulate the host’s blood glucose concentration and blood pressure [4],[5].Thus, they are classified as atypical pathogens.
3. Genome structure
The whole genome of Odoribacter splanchnicus' was sequenced in 2011 [3]. The O. splanchnicus genome contains a single 4,392,288 bp long chromosome with a G+C content of 43.4% [3]. There are 3,746 genes in total, including 3,672 protein-coding genes and 74 RNA genes [3]. Most protein-coding genes (61.2%) have been assigned to predicted basic cell functions, such as gene replication, transcription, translation, repair, cell membrane, metabolism, etc [3]. These genes, however, have yet to be fully described nor compared to any other species. There are also 175 pseudogenes identified, which are functionless genes resulting from multiple mutations [3]. Pseudogenes are also known to contain evolutionary history of the species, but further studies are needed to obtain such information [3].
4. Cell structure
Microbes within the Odoribacter sp. have a rod shape that tapers at the ends (commonly referred to as ‘fusiform’) [1]. They are Gram-negative, non-spore forming, and non-motile. Although several studies have identified and imaged different species under the Odoribacter sp. (Figure 1), a cellular model has yet to be developed [1],[3].
5. Metabolic processes
Members of the Odoribacter sp. are chemoorganotrophs that are strict anaerobes, as they lack catalases and oxidases which are required for aerobic metabolism [1],[3],[6]. Instead, they rely on the fermentation of carbohydrates as an energy source [2]. Most commonly, Odoribacter sp. are known for their butyrate-producing capabilities in the human gut. This process involves a butyrate kinase (Buk) pathway, which is common to several butyrate producing microbes [2]. Odoribacter splanchnicus, in particular, has a highly active pentose-phosphate pathway and is a large producer of hydrogen and hydrogen sulfide [3]. Odoribacter splanchnicus is also capable of tryptophan metabolism and hydrolyzing gelatin, however the mechanism underlying such metabolism has yet to be elucidated [6].
6. Ecology
A few strains of O. splanchnicus have been located in blood, while strains of O. denticanis have been traced in the gums of dogs [1],[2]. The majority of Odoribacter sp., however, predominantly presides in the gastrointestinal tract along and can be found in feces, appendixes, and peritoneal pusses of humans (2). O. splanchnicus has been shown to be culturable in lab, utilizing chopped meat media that contains carbohydrates at 37°C [3]. There is, however, little specification on its optimal growth conditions and no other Odoribacter sp. culturization has been cited in literature thus far [3]. Current research efforts are looking into how Odoribacter sp. function within the microbe community through their interactions with other gut microbiota and with their hosts.
7. Pathology
Odoribacter sp. are pathogenetic in domestic animals but often mutualistic and helpful to humans [1]. Maintaining the proper abundance of Odoribacter sp. in particular is crucial for keeping a healthy gut [7]. Loss of Odoribacter sp. results in reduced SCFA (short chain fatty acid) availability, leading to host inflammation, a symptom of the disease [2]. Additionally, Odoribacter sp. can help maintain lower systolic blood pressure in pregnant women through their production butyrate [2]. The presence of butyrate is known to reduce blood pressure and has been utilized in drugs for hypertension [8].
7. Key microorganisms
Include this section if your Wiki page focuses on a microbial process, rather than a specific taxon/group of organisms
8. Current Research
The most current research on Odoribacter sp. focuses on the correlation of its abundance with human health.
Odoribacter sp. & Blood Pressure
- A 2016 study composed of overweight and obese pregnant women at 16 weeks gestation showed that there was a correlation between the Odoribacter sp. population in the gut and systolic blood pressure, which is the blood pressure exerted on the arteries when the heart contracts to pump blood throughout the body [2]. Odoribacter sp. are well known for their production of butyrate via the Buk pathway [2]. The Buk gene encodes for butyrate kinase (a component of the Buk pathway), which catalyzes the process of butyryl-CoA to butyrate. Researchers found that both the abundance of Odoribacter sp. and the expression of Buk gene were negatively associated with the inflammatory marker plasminogen activator inhibitor-1 (PAI-1), a glycoprotein that positively correlates with the systolic blood pressure [2]. Therefore, the presence of Odoribacter sp. can be beneficial for maintaining healthy systolic blood pressure and can provide insight into novel methods for preventing hypertension or other high blood pressure related illnesses [2].
Odoribacter sp. & Type II Diabetes
- Another 2016 study on metformin, a treatment for Type II Diabetes, discovered a negative correlation between the abundance of the Bacteroidetes (the phylum of Odoribacter) and the glucose concentration in mice [5]. Furthermore, it has been observed that there is a significant decline in Odoribacter sp. in the offsprings of the mice that are exposed prenatally to metformin compared to the wild type offspring [5]. These findings suggest some association between Odoribacter sp. and diabetes, however, further studies should be conducted to explore the association.
9. References
Edited by [NAMES}, student of Jennifer Talbot for BI 311 General Microbiology, 2014, Boston University.
