Histoplasma capsulatum: Difference between revisions
| Line 33: | Line 33: | ||
==Cell metabolism== | ==Cell metabolism== | ||
Different nutrients and compounds are required for the growth of Histoplasma capsultum in different stages of its development. The nutritional requirements for the mycelia stage are simple; the organism can grow in a 25 °C with glucose as its sole source of carbon and ammonia as its source of nitrogen. During the yeast phase, H. capsulatum requires more complex compounds and a higher temperature environment. For the yeast phase, it first needs sulfhydryl-containing compounds for initiation of yeast development and cysteine or cystine along with certain growth factors such as biotin, thiamine, or thiotic acid in order to maintain the morphology. [1,10] Cysteine plays an important role in the morphogenesis of H. capsulatum. It allows the cell to perform respiration for both phases and during the transition. There are two terminal oxidase pathways for H. capsulatum: the cytochrome system which is blocked by cyanide and antimycin; the other, an unidentified alternative oxidase that is specifically blocked by salicylhydroxamic (SHAM). [1] | |||
Another nutrient that is important for the growth of Histoplasma capsulatum is Iron. Iron is essential in redox reaction related to its existence in reduced Fe2+ and oxidized Fe3+ states. For a pathogen to be successful, it must have countermechanisms for acquiring iron in the host microenvironment which it exists due to the limited amount of free soluble iron in an animal. There are a few approaches for the iron acquisition of H. capsulatum that have been studied. These strategies are: siderophores, acidic pH, reductive activities, and receptors for host iron-binding compounds. None of these approaches is unique because they can also be seen in other organisms; however, the coexistence of such plethora in a single microbe is distinctive. [4, 9] | |||
==Ecology== | ==Ecology== | ||
Revision as of 16:54, 27 August 2007
A Microbial Biorealm page on the genus Histoplasma capsulatum
Classification
Higher order taxa
Cellular Organisms; Eukaryota (Superkingdom); Fungi (Kingdom); Dikarya (Subkingdom); Ascomycota (Phylum); Pezizomycotina (Subphylum); Eurotiomycetes (Class); Eurotiomycetidae (Subclass); Onygenales (Order); Ajellomycetaceae (Family); Ajellomyces (Genus) [16]
Species
Species: Ajellomyces capsulata
Anamorph (an asexal reproductive stage): Histoplasma capsulatum [5]
Description and significance
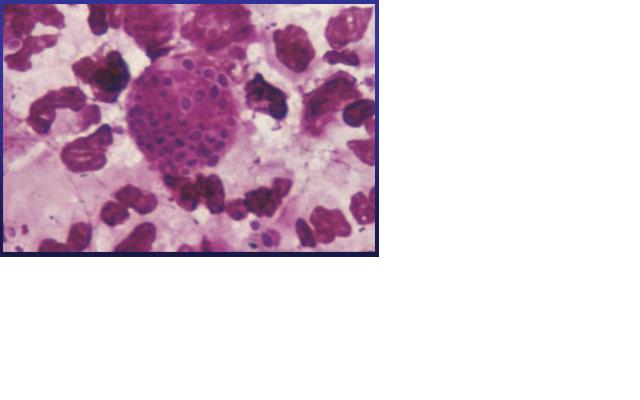
Histoplasma capsulatum is a fungal pathogen that can result in a wide range of clinical presentations, from asymptomatic through fatal infection. It usually causes a lung disease called Histoplamosis or Darling’s disease. It is called Darling’s disease because it was found by Samuel Darling in histopathologic specimens about a century ago. [2, 4]
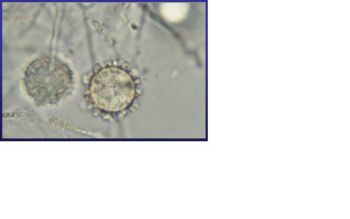
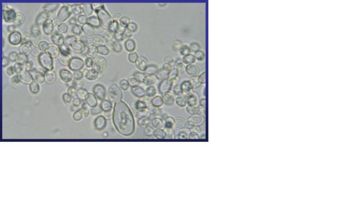
Histoplasma capsulatum is a biologically interesting inhabitant of soil and mammalian hosts, a clinically significant cause of respiratory and systemic infection, and an excellent fungal model of dimorphic cell development and facultative intracellular pathogenesis. [4] H. capsulatum is unique in its dimorphism which allows H. capsulatum to infect mammals by going through three significant development stages depends on the temperature shift from 25 °C to 37 °C. [1, 12] In the moist soil that is rich in bird or bat guano at temperature about 25 °C, H. capsulatum exists in a filamentous mycelia form. [4] However, when humans inhale H. capsulatum into their respiratory tracks, in order to replicate its DNA in the host at 37C, the pathogen has to be able to convert it’s tissue from one form to another. In this case, H. Capsulatum changed from fungi to yeast when it’s growing in the human bodies. [5, 16] H. capsulatum is thought to cause approximately 500,000 respiratory infections a year in the central river valleys in the Midwestern and south central United States. [2, 7, 11]
Histoplasma capsulatum reproduces when it is in the mold form of the fungus and is heterothallic. H. capsulatum is the anamorphic classification, while the teleomorph or perfect form is known as Ajellomyces capsulatus. Many approaches have been used to differentiate H. capsulatum strains. Three variants have been described based on host predilection, clinical presentation, geographic distribution, or serology: variant duboisii in Africa, variant farciminosum in horses, and variant capsulatum for the mijority isolates. [4, 5] The structure of H. capsulatum has not yet been studied with detail. There have been studies that focused on the polysaccharides composition of the cell walls. [5] The genome of H. capsulatum has also not yet been totally sequenced; however, certain strains of DNA have been studied. For examples, the G217B, G186A, and the Downs strain. [5] Many of the research centers are currently focusing on constructing the genome of H. capsulatum, such as University of Washington School of Medicine, and the Broad Institute. [8, 13] It will be useful to sequence the genome of H. capsulatum, because it can help answering many biological questions that scientists have. For example, the capacity to undergo the morphologic transition to the yeast phase, the different genes code for different functions necessary for survival, and the phase specific genes regulation. [1]
Genome structure
For Histoplasma capsulatum, functional genomic analysis preceded a genome sequence, demonstrating that genomics can be applied to organisms that have not been sequenced. [5, 9] However, some strains of DNA of H. capsulatum have been sequenced and studied. Most of the molecular studies of H. capsulatum were done with G217B, G186A, and the Downs strain. The G217B and the Downs strain are North American clinical isolates and G186A is a clinical isolate from Panama. [4, 5]
Originally H. capsulatum strains were classified into two types based on the polysaccharide composition of the cell walls. G217B belongs to type I, and it lacks α-(1,3)-glucan in its cell wall, while G186A belongs to type II because it has large amounts of α-(1,3)-glucan in its cell wall. This was interesting because for G186A variants that lack α-(1,3)-glucan are avirulent whereas G217B contains no α-(1,3)-glucan but is fully virulent. Therefore, the relationship between α-(1,3)-glucan and the H. capsulatum pathogenesis is relatable. [5]
Downs strain unlike G217B and G186A strains; it is avirulent in standard animal models. However, it is shown that Downs strain is a temperature-sensitive strain that has to do with patients who are probably immunocompromised and have Histoplasmosis. Studies of H. capsulatum Downs strain were done on AIDS patients and it showed close relationship between the patients’ immune system and the Downs strain. [1,5] Downs strain also plays a role in the sensitivity to the temperature change when H. capsulatum changes its forms from mycelium to yeast. It is shown that when the environment of the temperature is changing, H. capsulatum activates its “heat shock" (hs) genes during the induction of the yeast phase. These heat shock genes help H. capsulatum adapting new environment and allowing it to invade a human host. Downs strain heat shock protein peaked at 34 °C, and in strain G217B, the response was highest at 37 °C. [1, 9, 12] The comparison of the different strains of H. capsulatum genes aids the understanding the biology of H. capsulatum, perhaps future genome projects on H. capsulatum will be done and be able to help us increase our knowledge towards H. capsulatum.
Cell metabolism
Different nutrients and compounds are required for the growth of Histoplasma capsultum in different stages of its development. The nutritional requirements for the mycelia stage are simple; the organism can grow in a 25 °C with glucose as its sole source of carbon and ammonia as its source of nitrogen. During the yeast phase, H. capsulatum requires more complex compounds and a higher temperature environment. For the yeast phase, it first needs sulfhydryl-containing compounds for initiation of yeast development and cysteine or cystine along with certain growth factors such as biotin, thiamine, or thiotic acid in order to maintain the morphology. [1,10] Cysteine plays an important role in the morphogenesis of H. capsulatum. It allows the cell to perform respiration for both phases and during the transition. There are two terminal oxidase pathways for H. capsulatum: the cytochrome system which is blocked by cyanide and antimycin; the other, an unidentified alternative oxidase that is specifically blocked by salicylhydroxamic (SHAM). [1] Another nutrient that is important for the growth of Histoplasma capsulatum is Iron. Iron is essential in redox reaction related to its existence in reduced Fe2+ and oxidized Fe3+ states. For a pathogen to be successful, it must have countermechanisms for acquiring iron in the host microenvironment which it exists due to the limited amount of free soluble iron in an animal. There are a few approaches for the iron acquisition of H. capsulatum that have been studied. These strategies are: siderophores, acidic pH, reductive activities, and receptors for host iron-binding compounds. None of these approaches is unique because they can also be seen in other organisms; however, the coexistence of such plethora in a single microbe is distinctive. [4, 9]
Ecology
Describe any interactions with other organisms (included eukaryotes), contributions to the environment, effect on environment, etc.
Pathology
How does this organism cause disease? Human, animal, plant hosts? Virulence factors, as well as patient symptoms.
Application to Biotechnology
Does this organism produce any useful compounds or enzymes? What are they and how are they used?
Current Research
Enter summaries of the most recent research here--at least three required
References
1. Bossche, Hugo, Frank Odds, and David Kerridge. Dimorphic Fungi in Biology and Medicine. 1st ed. New York and London: Plenum Press, 1993
2. Chang, Ryan. "Histoplasmosis." 19 Sep 2005. emedicine. 25 Aug 2007 <http://www.emedicine.com/MED/topic1021.htm>.
3. Hayward, Chris A. "Microorganisms." Encyclopedia of Life Sciences 20062006 1-13. <http://www.mrw.interscience.wiley.com/emrw/9780470015902/els/article/a0000460/current/pdf>.
4. Heitman, Joseph, Scott G. Filler, Aaron P. Mitchell, and John E. Edwards, Jr.Molecular Principles of Fungal Pathogenesis. 1st ed. New York: ASM Press, 2006. (Heitman et al. 321-331)
5. Heitman, Joseph, Scott G. Filler, Aaron P. Mitchell, and John E. Edwards, Jr.Molecular Principles of Fungal Pathogenesis. 1st ed. New York: ASM Press, 2006. (Heitman et al. 611-626)
6. "Histoplasmosis." Histoplasmosis Resource Guide. Mar 2007. National Eye Institute. 25 Aug 2007 <http://www.nei.nih.gov/health/histoplasmosis/index.asp>.
7. "Histoplasmosis." Centers for Disease Control and Prevention. 12 Oct 2005. Centers for Disease Control and Prevention. 25 Aug 2007 <http://www.cdc.gov/ncidod/dbmd/diseaseinfo/histoplasmosis_g.htm>.
8. "Histoplasma capsulatum Database." BROAD INSTITUTE. 20 Aug 2007. BROAD INSTITUTE. 20 Aug 2007 <http://www.broad.mit.edu/annotation/genome/histoplasma_capsulatum/Home.html>.
9. Hwang, Lena, Davina Hocking-Murray, Adam K. Bahrami, Margareta Andersson, and Jasper Rine. "Identifying Phase-specific Genes in the Fungal Pathogen Histoplasma capsulatum Using Genomic Shotgun Microarray." Molecular Biology of the Cell June 2003 2314-2326. .
10. Johnson, Clayton, Jonathan T. Prigge, Aaron D. Warren, and Joan E. McEwen. "Characterization of an alternative oxidase activity of Histoplasma Capsulatum." Yeast 2003 381-388. .
11. Kendrick, Bryce. "Fungi: Ecological Importane and Impact on Humans." Encyclopedia of Life Sciences 20012001 1-5. <http://www.mrw.interscience.wiley.com/emrw/9780470015902/els/article/a0000369/current/pdf>.
12. Kobayashi, George S. "Mycoses." Encyclopedia of Life Sciences 20012001 1-6. <http://www.mrw.interscience.wiley.com/emrw/9780470015902/els/article/a0000371/current/pdf>.
13. Kobayashi, George S.. "Molecular Basis of Adaptation in Histoplasma capsulatum ." Washington University School of Medicine. 25 Aug 2007 <http://research.medicine.wustl.edu/OCFR/Research.nsf/Abstracts/64ABA9996B650C50862567ED00029E69?OpenDocument&VW=Infectious+Diseases>.
14. Lai, Chung-Hsu, Chun-Kai Huang, Chuen Chin, Ya-Ting Yang, and Hsiu-Fang Lin. "Indigenous Case of Disseminated Histoplasmosis, Taiwan." Emerging Infectious Diseases Jan 2007 127-129. .
15. Maubon, Daniele, Stephane Simon, and Christine Aznar. "Histoplasmosis diagnosis using a polymerase chain reaction method. Application on human samples in French Guiana, South America." 04 Aug 2007
16. Mitchell, Thomas G. "Fungal Pathogens of Human." Encyclopedia of Life Science 20022002 1-15. <http://www.mrw.interscience.wiley.com/emrw/9780470015902/els/article/a0000359/current/pdf?hd=All%2Chistoplasma&hd=All%2Ccapsulatum>.
17. National Center for Biotechnology Information (NCBI) Taxonomy Browser. Taxonomy ID:5037
18. Nittler, M.Paige, Davina Hocking-Murray, Catherine K. Foo, and Anita Sil. "Identification of Histoplasma capsulatum Transcripts Induced in Response to Reactive Nitrogen Species." Molecular Biology of the Cell Oct 2005 4792-4813. .
19. Rappleye, C. A., Eissenberg, L. G. & Goldman, W. E. Histoplasma capsulatum (1,3)-glucan blocks innate immune recognition by the -glucan receptor. Proc. Natl Acad. Sci. USA 23, 1366–1370 (2007)
Image Source: 20. Scientific Institute of Public Health, Belgium, http://www.iph.fgov.be/
Edited by Shen-Yin (Mandy) Hung of Rachel Larsen