Zaire ebolavirus
Classification
Higher order taxa
Group: Group; V ((-)ssRNA) Order; Mononegavirales Family; Filoviridae Genus; ebolavirus Species; Zaire
Species
|
NCBI Taxonomy: [7] |
Zaire ebolavirus
Description and significance
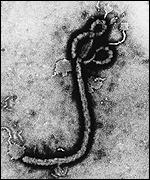
The appearance of this negative sense single stranded RNA (nonsegmented) virus is filamentous in structure(NIH, 2010)[8]. This virus was discovered in the Democratic Republic of Congo and is a species of the Genus ebolavirus. The Genus ebolavirus contains five different species. The five species are: Sudan ebolavirus (SEBOV), Reston ebolavirus (REBOV),Bundibugyo ebolavirus (BEBOV), Ivory Coast ebolavirus (CIEBOV), and Zaire ebolavirus (ZEBOV)(NIH, 2010)[9]. They were all discovered around 1976 with only one strain that is not pathegenic in humans which is the Reston ebolavirus. The filoviruses can cause up to a 90% mortality rate in humans as well(NIH, 2010)[10]. The virus is not considered to be living due to the fact that without a host the virus will die, although this topic is highly controversial.
Genome structure
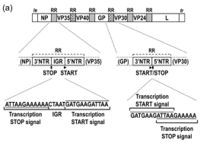
The complete genome is 18,959 base pairs which makes up a linear strand of RNA. This is a single stranded, nonfragmented, negative sense RNA, which resides in Group V of the Baltimore classifcation system(Sanchez et al., 1993)[11]. This class of viruses needs a RNA-dependent RNA polymerase to transcribe the negative sense strand in to a positive sense strand. This can only be conducted by a viral RNA-dependent RNA polymerase(Volchkov et al, 1999)[12].
The Zaire Ebola virus consists of 7 linearly arranged genes. The order of the genes is as follows: 3'-NP-VP35-VP40-GP-VP30-VP24-L. The transcriptional start and stop signals contain the sequence 3′-UAAUU(Volchkov et al, 1999)[13]. Three base intergenic sequences are present between the NP and VP35 genes (3′-GAU) and VP40 and GP genes (3′-AGO, and a large intergenic sequence of 142 bases separates the VP30 and VP24 genes(Volchkov et al, 1999)[14]. Stem-and-loop structures were identified in the 5I end of the leading strand. Alignment of predicted amino acid sequences showed that the structural proteins of Ebolavirus and Marburgvirus contain large regions of homology(Volchkov et al, 1999)[15].
Cell structure and metabolism
VP30, VP35, nucleoprotein, and a polymerase protein [L] are the 4 virion structural proteins. VP40, glycoprotein [GP], and VP24 are the 3 membrane-associated proteins(Sanchez et al., 1996)[16]. The surface GP is encoded in 2 open readingframes (ORFI and ORFII). ORF I (amino-terminal) encodes for a small (50-70 kd), soluble, nonstructural secretory glycoprotein (sGP) that is produced in large quantities early in Ebola infection(Sanchez et al, 1996)[17].
The sGP binds to neutrophil CD16b, a neutrophil-specific Fc g receptor III, and inhibits early neutrophil activation. This could account for the lymphopenia associated with an Ebola infection. According to this research sGP is likely the key factor in preventing a successful immune response from the host(Sanchez et al., 1996)[18].
However, more recent research has proven that neutrophils do not express a specific receptor for Ebolavirus sGP. It is unlikely that sGP plays a role in the Ebolavirus pathogenesis through interfering with the innate immunity by targeting neutrophils, so the exact metabolism of the Ebolavirus is still unknown(Sanchez et al., 1996)[19].
Ecology
Zaire ebolavirus RNA has been discovered within the cells of rodents and shrews that occupy niches in forest areas. This concludes that reservoirs of this virus are found in Central African Republic inside the rodent's internal organs(Feldmann et al., 2004)[20]. It has started a recent epidemic in the Republic of Congo, originally called Zaire, which has lead to a sudden death in mammals and humans. This zoonotic pathogen can be transmitted from animals to other uninfected animals and jump to invade humans(Feldmann et al., 2004)[21].
Zaire ebolavirus is a parasite to humans; therefore, the symbiosis is a parasitism relationship. The virus benefits and the host is negatively impacted during their interaction(Feldmann et al., 2004)[22] This Zaire ebolavirus is found within mammals that consume the natural vegetation of the forest environment of Africa. This negatively impacts wildlife like apes, chimpanzees, and bats. Once transferred to human, this becomes a devastation to public health(Feldmann et al., 2004)[23].

In a recent study conducted in the rural areas of Gabon, a high rate of Zaire ebolavirus immunity(humoral and cellular) was discovered within the forest tribes of that region(Becquart et al., 2010)[24]. There were 4,349 people involved in this study which encompassed, 220 randomly selected villages. The prevalence of anti-ZEBOV lgG in an indiviual was what determined the immunity for each community. For the people who lived in the forest, they had the highest rate of immunity (19.4%) compared to people who lived by the lake, on a savannah, or on the grasslands(Becquart et al., 2010)[25]. The next highest rate of immunity came from the people who lived on the grasslands which was 12.4%. The scientists believe that the high rate of immunity discovered in the forest communities comes from close contact of human exposure, bat saliva, and fruit contaminated by the virus(Becquart et al., 2010)[26].
Pathology
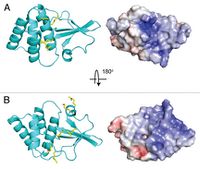
Out of any species of the Ebolavirus, the Zaire species is by far the most lethal. The fatality rate can reach up to 90% in some cases and death occurs within days of the initial infection(Leung et al., 2010)[27]. The symptoms include: shock, fever, and internal coagulation which can determine how rapidly the virus is replicated. The host is usually human, however the virus can be spread to primates. There is a protein called the Ebola viral protein 35 (VP35) which is a cofactor for the Ebola RNA polymerase and this complex is needed for viral production(Leung et al., 2010)[28]. The virulence factor for the Zaire ebolavirus is when VP35 binds to double strand RNA (dsRNA). Recently, an end-cap was discovered for the VP35 protein, which are hydrophobic molecules attached to the end tail. This end-cap actually helps the VP35 protein bind to dsRNA(Leung et al., 2010)[29]. This end-cap can also mimic the espression of RlG-l-Like receptors (RLR) which is one of the reasons the virus is so successful in taking down the hosts immune system. The VP35 protien is a great target when treating the Ebolavirus[30]. There have been two different methods tested which are by using nucleic acids and siRNA molecules.
When the immune system first reacts to the Zaire ebolavirus, a large quantiy of chemokines, cytokines, and growth factors are produced by the body. Also, it was noted that there is lymphocyte apoptosis due to the apparent loss of peripheral CD4 and CD8 lymphocytes(Warren et al., 2010)[31]. Thus, it is easy for the virus to take over other cells if the lymphocyte production is low. In another study using a mouse strain of the Zaire ebolavirus, the treatment using FGI-103 proved to be an efficient way to protect the mice(Warren et al., 2010)[32]. For the 10 control mice, the typical 90% mortality rate was observed. However, for the 10 mice that received 10mg/kg of FGI-103 after 1,000 PFU ZEBOV, they were protected and were healthy after 10 days(Warren et al., 2010)[33]. This study is promising in finding a treatment for Zaire ebolavirus while also keeping the mortality rate down.
Current Research
Human Fatal Zaire Ebola Virus Infection Is Associated with an Aberrant Innate Immunity and with Massive Lymphocyte Apoptosis
The Zaire ebolavirus is a deadly infectious pathogen that can suppress the immune system. It has a case-fatality rate of 90% in patients who are infected due to a lethal hemorrhagic fever syndrome. This Zaire ebolavirus is confirmed infected patients blood by the presence of a specific non-self antigen. This virus attacks the adaptive immune response by decreasing the amount of B lymphocytes and T lymphocytes present(Waquier et al., 2010)[34]. These B cells and T-cells such as CD4 and CD8 lymphocytes are necessary to produce cytokines as immune mediators. Patients who die within a couple days show a decrease in of B and T cells numbers due to apoptosis, cell programmed death. T cell are necessary to be activated to kill off virus infected cells. Researchers have been trying to make a vaccine to cure this disease. They have performed in vitro studies with mice by injecting them with a strain and Zaire ebolavirus and performing assays on cytokines and detecting by fluorescence for final concentrations. These have provided consistent results for further studies. (Wauquier et al., 2010)[35].
The Tyro3 Receptor Kinase Axl Enhances Macropinocytosis of Zaire Ebolavirus
A TAM family member, AxI, is connected to the plasma membrane and is what allows the Zaire ebolavirus glycoprotein to enter some cells, but not all cells(Hunt et al., 2011)[36]. Also, how the AxI specifically works in order to allow viruses to enter the cell is not completely known, so this study examined all entries in to the cell that ZEBOV could use(Hunt et al., 2011)[37]. The scientists used interfering RNA in order to discover the pathways that ZEBOV used in order to enter the cell. The main pathway that was discovered for ZEBOV to enter the cell was through macropinocytosis. Also, the RNAi was used to study how AxI actually stimulates macropinocytosis and uptake of ZEBOV. It was concluded that AxI is necessary in order for ZEBOV transduction and also Zaire ebolavirus glycoprotein-dependent virions uptake. This experiment could prove to be very useful in inhibiting ZEBOV from enter the cell, if all of the exact entries are found(Hunt et al., 2011)[38].
Demonstration of Cross-Protective Vaccine Immunity against an Emerging Pathogenic Ebolavirus Species
A recent study has allowed us to delve into the expansion of our vaccines to branch into the other species within the Zaire ebolavirus. A study was conducted in which current non-human primate vaccine, specifically EBOV GP DNA, was combined with rAd5 boost immunization (Hensley, L. E., et al 2010)[39]. The DNA vaccine uses a plasmid vector of glycoproteins, GP (Z) and GP (S/G) while the rAd5-GP is a strong antibody and generates CD4+ T cells and CD8+ T cells(Hensley, L. E., et al 2010)[40]. These cells are important for long term memory for the vaccine and enhance CD8+ T cell functions. This allows the vaccines to be boosted “long term” after a time period of one year. The DNA/rAd5 vaccine protects non-human primates against the BEBOV species, although there is no specific vaccine targeting this species. The vaccine works to attack the homologous structures in BEBOV, similar to those found in SEBOV and ZEBOV. This perpetuates cross-protective immunity when animals were injected with the BEBOV virus. There is a conservation of structures between species despite their divergence (Hensley, L. E., et al 2010)[41]. This study allows us to gain insight into homologous structures and allow us to put out focus on these types of advances. Since the Zaire ebolavirus can be used as a weapon of bioterrorism developing a vaccine for all species is necessary and prudent.[42]
Cool Factor
Zaire ebolavirus can potentially be used as a bioweapon due to its lethal effect on humans. There is a serious movement conducted by scientists in order to create a treatment for this virus, in case it were to ever be used as a weapon. As of right now, there is no vaccine in order to treat a ZEBOV infection for humans but there has been some progression of creating a vaccinef or animal models(Sanchez et al., 1996)[43]. The virus has also been the star in many fictional works, such as The Hot Zone by Richard Preston and the muse behind movies like, "Outbreak".
References
[44] Becquart, P., Wauquier, N., Mahlakõiv, T., and Nkoghe, D. "High Prevalence of Both Humoral and Cellular Immunity to Zaire ebolavirus among Rural Populations in Gabon", 'PLoS One'. 2010. Volume 5.
[45]."Electron Micrograph of Zaire ebolavirus." Photograph. BBC. First Last. London: BBC NEws, 1997. Web. 24 Oct 2011.
[46] Feldmann H, Wahl-Jensen V, Jones S, and Stroher U (2004). Ebola Virus Ecology: A Continuing Mystery.Trends in Microbiology.Vol 12(10):433-437.
[47]Hensley, L. E., Mulangu, S., Asiedu, C., Johnson, J., Honko, A. N., Stanley, D., & ... Sullivan, N. J. (2010). Demonstration of Cross-Protective Vaccine Immunity against an Emerging Pathogenic Ebolavirus Species. Plos Pathogens, 6(5), 1-9. doi:10.1371/journal.ppat.1000904
[48] Leung, D. W., Prins, K. C., Basler, C. F., & Amarasinghe, G. K. (2010). Ebolavirus vp35 is multifunctional virulence factor. Virulence, 1(6), 526-531. doi: 10.4161/viru.1.6.12984
[49] NCBI Taxonomy Zaire ebolavirus
[50] National Institutes of Health. "Botulinum Neurotoxin, B. Anthracis and Variola Virus" 'National Academies Press'. 2010
[51] Olejnik, J., Ryabchikova, E., Corley, R.B, and Mühlberger, E. "Intracellular Events and Cell Fate in Filovirus Infection", 'Viruses'. 2011. Volume 8. p.1501-1531
[52] Sanchez, Anthony, Kiley, Michael P, Holloway, Brian P, Auperin, David D. (1993). Sequence analysis of the Ebola virus genome: organization, genetic elements, and comparison with the genome of Marburg virus. Virus Research Vol. 29(3): 215-240.
[53] Sanchez A, Trappier SG, Mahy BW, Peters CJ, Nichol ST (1996). The virion glycoproteins of Ebola viruses are encoded in two reading frames and are expressed through transcriptional editing. Proc Natl Acad Sci U S A. Vol. 93(8):3602-3607.
[54] Sui, Jianhua, Marasco, Wayne A. (2002). Evidence against Ebola Virus sGP Binding to Human Neutrophils by a Specific Receptor. Virology. Vol. 303(1): 9-14.
[55] Volchkov,V.E., Volchkova,V.A., Chepurnov,A.A., Blinov,V.M., Dolnik,O., Netesov,S.V. and Feldmann,H. "'Zaire ebolavirus', 'Characterization of the L gene and 5' trailer region of Ebola virus'". 'Journal of General Virology'. 1999. Volume 80. p.355-362
[56] Wauquier N., Becquart P, Padilla C, Baize S, Leroy E (2010) Human Fatal Zaire Ebola Virus Infection Is Associated with an Aberrant Innate Immunity and with Massive Lymphocyte Apoptosis. Public Library of Science Vol. 4(10):837-847
[57] Warren, T.K, Warfield, K.L, Wells, J., and Enterlein, S. "Antiviral Activity of a Small-Molecule Inhibitor of Filovirus Infection", 'Antimicrobial Agents and Chemotherapy'. 2010. Volume 54. p.2152-2159
Edited by student of Iris Keren
