Acidobacteria capsulatum: Difference between revisions
No edit summary |
No edit summary |
||
| Line 12: | Line 12: | ||
==Description and significance== | ==Description and significance== | ||
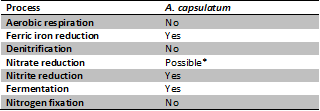
[[File:Table_A_capsulatum_Process.png| | [[File:Table_A_capsulatum_Process.png|500px|thumb|left| '''Fig 1'''Summary of known major metabolic processes absent or present within ''A. capsulatum''.*Presence of some of the genes required for pathway (2)]] | ||
''Acidobacteria capsulatum'' (''A. capsulatum'') is still a relatively new species since its official classification in 1991 but progress has been made towards elucidating the properties and ecological significance of the species. ''A. capsulatum'' are gram-negative, acidophilic, heterotrophic, facultatively anaerobic and rod shaped bacteria ('''1'''). | ''Acidobacteria capsulatum'' (''A. capsulatum'') is still a relatively new species since its official classification in 1991 but progress has been made towards elucidating the properties and ecological significance of the species. ''A. capsulatum'' are gram-negative, acidophilic, heterotrophic, facultatively anaerobic and rod shaped bacteria ('''1'''). | ||
| Line 21: | Line 21: | ||
==Ecology== | ==Ecology== | ||
[[File:Acid_mine.gif| | [[File:Acid_mine.gif|500px|thumb|right|Acid mine drainage ]] | ||
''A. capsulatum'', though first isolated from [http://en.wikipedia.org/wiki/Acid_mine_drainage acidic drainage] ('''1'''), are presumed to be widely distributed in soils as evidenced by the detection of their phylum’s 16s rRNA sequences across a diverse range of environmental samples ('''4'''). Soil samples from Australian forest and Arizona indicated that ''A. capsulatum '' was present in the microbial communities. Additionally '' A. capsulatum '' also extends its presence to [http://en.wikipedia.org/wiki/Rhizosphere rhizosphere soils], the interface between the plant root and microbial community. Here, members of the ''Acidobacteria '' phylum including '' A. capsulatum '' were found to have represented a majority of the 16s rRNA sequences found there ('''5'''). ''A. capsulatum '' then may play a role in the geochemical cycles within these soils. | ''A. capsulatum'', though first isolated from [http://en.wikipedia.org/wiki/Acid_mine_drainage acidic drainage] ('''1'''), are presumed to be widely distributed in soils as evidenced by the detection of their phylum’s 16s rRNA sequences across a diverse range of environmental samples ('''4'''). Soil samples from Australian forest and Arizona indicated that ''A. capsulatum '' was present in the microbial communities. Additionally '' A. capsulatum '' also extends its presence to [http://en.wikipedia.org/wiki/Rhizosphere rhizosphere soils], the interface between the plant root and microbial community. Here, members of the ''Acidobacteria '' phylum including '' A. capsulatum '' were found to have represented a majority of the 16s rRNA sequences found there ('''5'''). ''A. capsulatum '' then may play a role in the geochemical cycles within these soils. | ||
| Line 40: | Line 40: | ||
==Genome Structure== | ==Genome Structure== | ||
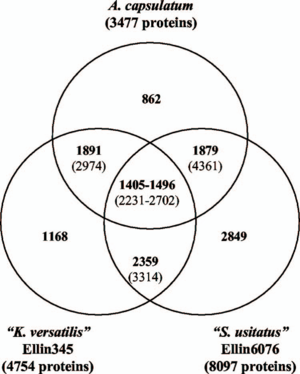
[[File:Three_Genome.png| | [[File:Three_Genome.png|300px|thumb|left|'''Fig 2''' Shared genes between ''A. capsulatum, K. versatilis'' and ''S. usitatus.'' ('''2''') ]] | ||
''A. capsulatum'' has a relatively small genome size of approximately 4.12 million base pairs, whereas other Acidobacteria species such as ''K. versatilis'' and ''S. usitatus'' have about 5.65 and 10 million base pairs respectively. The size of ''A. capsulatum''’s genome is attributed to it being more specialized in comparison to other members of the phylum, being able to degrade less substrate than the others. Comparative genome analysis between the three bacterial species ('''Fig 2''') showed most of the shared genes encoded for conserved cell function. Genes that were different represented some difference in pathways and substrate preference. For example, unlike the other two, ''A. capsulatum'' does not encode cellulases from the family 1 Beta-glucosidases, suggesting the lack of cellulose degradation for ''A. capsulatum''. Interestingly, ''A. capsulatum'' has the genes required for cellulose synthesis ('''2'''). | ''A. capsulatum'' has a relatively small genome size of approximately 4.12 million base pairs, whereas other Acidobacteria species such as ''K. versatilis'' and ''S. usitatus'' have about 5.65 and 10 million base pairs respectively. The size of ''A. capsulatum''’s genome is attributed to it being more specialized in comparison to other members of the phylum, being able to degrade less substrate than the others. Comparative genome analysis between the three bacterial species ('''Fig 2''') showed most of the shared genes encoded for conserved cell function. Genes that were different represented some difference in pathways and substrate preference. For example, unlike the other two, ''A. capsulatum'' does not encode cellulases from the family 1 Beta-glucosidases, suggesting the lack of cellulose degradation for ''A. capsulatum''. Interestingly, ''A. capsulatum'' has the genes required for cellulose synthesis ('''2'''). | ||
Revision as of 05:23, 27 December 2012
Classification
Higher order taxa
Bacteria; Fibrobacteres/Acidobacteria group; Acidobacteria; Acidobacteriia; Acidobacteriales; Acidobacteriaceae; Acidobacterium
Species
Acidobacterium capsulatum
|
NCBI: Taxonomy |
Description and significance
Acidobacteria capsulatum (A. capsulatum) is still a relatively new species since its official classification in 1991 but progress has been made towards elucidating the properties and ecological significance of the species. A. capsulatum are gram-negative, acidophilic, heterotrophic, facultatively anaerobic and rod shaped bacteria (1).
A. capsulatum are known to possess flagella that allow motility within the environment (1). The flagella themselves are peritrichous or found all over the surface of the cell. Another characteristic of this organism is the presence of high amounts of exopolysaccharide coating the cells from soil isolates (2). Exopolysaccharide’s function falls into four broads categories: protection, adhesion, an adaption response or cell-cell interactions (3). This contributes to the competitiveness of A. capsulatum, for example, having increased adhesion can allow the bacterium to acquire nutrients more readily from the environment.
Members of this phylum are presumably ubiquitous in soil environments and A. capsulatum could potentially play a major role in the fluxes of carbon,nitrogen and iron across microbial communities due to its metabolic processes (Fig 1) and distribution in soils.
Ecology
A. capsulatum, though first isolated from acidic drainage (1), are presumed to be widely distributed in soils as evidenced by the detection of their phylum’s 16s rRNA sequences across a diverse range of environmental samples (4). Soil samples from Australian forest and Arizona indicated that A. capsulatum was present in the microbial communities. Additionally A. capsulatum also extends its presence to rhizosphere soils, the interface between the plant root and microbial community. Here, members of the Acidobacteria phylum including A. capsulatum were found to have represented a majority of the 16s rRNA sequences found there (5). A. capsulatum then may play a role in the geochemical cycles within these soils.
A. capsulatum prefer to exist in conditions where the soil or the microenvironments within it provide a pH range of 3.0 – 6.0 (6). In addition, they are frequent and sometimes dominant in iron-rich environments such as abandoned mines (2). This can be attributed to their mode of respiration, iron-reduction, which can play a significant role in iron cycling in these environments.
Metabolism
A. capsulatum are facultative anaerobes (7), though previously they were described to be aerobic when they were first discovered (1). When oxygen concentrations are low or depleted, A. capsulatum are able to use dissimilatory ferric iron reduction (6). As ferric iron exists more favourably in low pH levels, the tolerance to acidity appears adaptive for these microbes (Fig 2). The frequency of A. capsulatum in soils may reflect the important role it plays in the iron cycle and microbial community by providing ferrous iron, produced from ferric iron reduction, for nutrient uptake in organisms.
A. capsulatum has the genes required for cellulose synthesis, allowing the potential production of bacterial cellulose. This is unique for A. capsulatum as the genes are rare in non-proteobacterial genomes. Bacterial cellulose biofilm is hypothesized to be able to promote and facilitate adherence to ferric-iron substrates (8), which can be quite useful in nutrient starved environments. Furthermore, the benefits of cellulose biofilms also include protection from desiccation or drying out and physical support. A. capsulatum then would seem to be equipped to survive in stressful environments.
A. capsulatum may express an addiction module, or a toxin-antitoxin system (2). Toxins produce by this system are known to inhibit DNA synthesis and relatedly, protein synthesis (9). This system allows A. capsulatum to adapt to environments where nutrients are scarce by reducing its requirement for energy.
A. capsulatum may be capable of denitrification, as nitrate reductase genes analogous to those found in cyanobacteria were present within A. capsulatum’s genome (2). They are however, capable of nitrite reduction.
The diverse modes of respiration and survival mechanisms in the A. capsulatum’s repertoire highlights its ability to adapt to a range of environments and could explain its abundance in soils, where conditions can change rapidly and the scarcity of terminal electron acceptors limit cell growth.
Genome Structure
A. capsulatum has a relatively small genome size of approximately 4.12 million base pairs, whereas other Acidobacteria species such as K. versatilis and S. usitatus have about 5.65 and 10 million base pairs respectively. The size of A. capsulatum’s genome is attributed to it being more specialized in comparison to other members of the phylum, being able to degrade less substrate than the others. Comparative genome analysis between the three bacterial species (Fig 2) showed most of the shared genes encoded for conserved cell function. Genes that were different represented some difference in pathways and substrate preference. For example, unlike the other two, A. capsulatum does not encode cellulases from the family 1 Beta-glucosidases, suggesting the lack of cellulose degradation for A. capsulatum. Interestingly, A. capsulatum has the genes required for cellulose synthesis (2).
Much of the proteins encoded by A. capsulatum’s genome are closely related to proteins in the phylum Proteobacteria (2). In addition, phylogenetic trees using 16s rRNA markers also show Acidobacteria and Proteobacteria being closely related compared to other phylums (10). A. capsulatum and other Acidobacteria tend to co-occur with members of the Proteobacteria phylum; it is possible for lateral gene transfer and co-evolution to occur between the two and this may explain the similarities in genes.
Current Research
There is research being conducted by researchers at Fort Lewis College, investigating A. capsulatum’s complete metabolic pathway and enzymes (11).
Relatedly, future research as suggested by Ward et al. should be directed towards understanding how A. capsulatum functions in a living system or ecosystem in order to elucidate gene function (2).
References
1. Kishimoto, N., Kosako, Y. and Tano, T. 1991. Acidobacterium capsulatum gen. nov., sp. nov.: An Acidophilic Chemoorganotrophic Bacterium Containing Menaquinone from Acidic Mineral Environment. Current Microbiology. 22:1–7.
2. Ward, N. L, et al. 2009. Lifestyles of These Microorganisms in Soils Acidobacteria Provide Insight into the Three Genomes from the Phylum. Appl. Environ. Microbiol. 75:2046.
3. Weiner, R., Langille ,S., and Quintero, E. 1995. Structure, function and immunochemistry of bacterial exopolysaccharides. J. Ind. Microbiol. 15:339–346.
4. Quaiser A, Ochsenreiter T, Lanz C, et al. 2003. Acidobacteria form a coherent but highly diverse group within the bacterial domain: evidence from environmental genomics. Mol. Microbiol. 50: 563–75.
5. Lee S. H., Ka J. O., and Cho J. C. 2008. Members of the phylum Acidobacteria are dominant and metabolically active in rhizosphere soil. FEMS Microbiol. Lett. 285: 263–269.
6. Blothe, M., Akob, D. M., Kostka, J. E., Goschel, K., Drake, H. L. & Kusel, K. 2008. pH gradient-induced heterogeneity of Fe(III)-reducing microorganisms in coal mining-associated lake sediments. Appl. Environ. Microbiol. 74:1019–1029.
7. Pankratov, T. A., Kirsanova, L. A., Kaparullina, E. N., Kevbrin, V. V. and Dedysh, S. N. 2012. Telmatobacter bradus gen. nov., sp. nov., a cellulolytic facultative anaerobe from subdivision 1 of the Acidobacteria, and emended description of Acidobacterium capsulatum Kishimoto et al. 1991. Int. J. Syst. Evol. Microbiol. 62:430–437.
8. Kammler, M., C. Schon, and K. Hantke. 1993. Characterization of the ferrous iron uptake system of Escherichia coli. J. Bacteriol. 175:6212–6219.
9. Chang, D. E., D. J. Smalley, and T. Conway. 2002. Gene expression profiling of Escherichia coli growth transitions: an expanded stringent response model. Mol. Microbiol. 45:289–306.
10. Kuske, C.R., Barns, S.M., and Busch, J.D. 1997. Diverse uncultivated bacterial groups from soils of the arid southwestern United States that are present in many geographic regions. Appl. Environ. Microbiol. 63: 3614–3621.
11. http://www.fortlewis.edu/chemistry/DepartmentalNewsRecognitions.aspx