Agrobacterium tumefaciens
A Microbial Biorealm page on the genus Agrobacterium tumefaciens

Classification
Higher order taxa
Bacteria (Domain); Proteobacteria (Phylum); Alphaproteobacteria (Class); Rhizobiales (Order); Rhizobiaceae (Family); Agrobacterium (Genus)
Species
|
NCBI: Taxonomy |
Agrobacterium tumefaciens
Description and significance

Agrobacterium tumefaciens is a gram-negative rod-shaped bacteria (Jin, S.) closely related to nitrogen-fixing bacteria which dwell at root nodules in legumes. Unlike most other soil-dwelling bacteria, it infects the roots of plants to cause Crown Gall Disease (Jin, S.). In the wild A. tumefaciens targets dicots, and causes economic damage to plants with agricultural importance such as walnuts, tomatoes and roses. Various remediation methods, including utilization of a strain of closely related bacteria (Agrobacterium radiobacter) (Ryder, M. H.) controls and limits its damage, but it is also useful as a genetic engineering tool in plants. It is famous for taking advantage of its host by injecting DNA derived from its Ti (tumor inducing) plasmid into its host, causing the plant to create galls which excrete opines that the bacteria use as an energy source. However, scientists have exploited this ability of this bacteria to put DNA into its host to create transgenic plants. A. tumefaciens have emerged as an important molecular tool for manipulating plants and creating genetically modified crops for research and agriculture. Because of its importance in the laboratory, a complete genome of A. tumefaciens strain C58 was published in 2001 (Goodner, B.).
Genome structure
A. tumefaciens is an unusual bacteria because is one of the few that has both a linear and a circular chromosome. Its genome has a total of 5.7 million base-pairs, with 2.8 million residing on its circular chromosome and 2.1 million residing on its linear chromosome (Goodner, B). Most of the genes essential for its survival are located on the circular chromosome, although through evolution some essential genes have migrated to the linear chromosome. Based on sequence analysis, it was determined that the linear chromosome was derived from a plasmid that was transformed into the bacteria a long time ago (Goodner, B).
The ends of the linear chromosomes are protected by a telomere that forms a covalently closed hairpin, like in other bacteria which contain a linear chromosome (Goodner, B). In addition to the two chromosomes, strain C58 also contain two plasmids, pTiC58 (generically called Ti) and pAtC58 (also called the "cryptic plasmid"). pTiC58 contains genes necessary for its pathogenicity against plants (Goodner, B.), including the T-DNA which is injected into the plant and causes it to produce opines, along with accessory proteins which helps the T-DNA enter and transform the plant cell into a tumor cell. It is believed that pAtC58 contains genes essential for opine catabolism (Vaudequin-Dransart, V.), or its ability to use opines as an energy source, which is important for its lifestyle as a pathogen.
Pathology
A. tumefaciens infects plants usually through an open wound. It seeks out phenolic compounds which spills from a plant wound and chemotactically moves toward its source. Once it enters its plant host, it injects a section of its DNA called the T-DNA which is derived from its Ti (tumor inducing) plasmid into its host (Moore, L. W.). The T-DNA is then integrated into the plant's genome, and has two effects on the plant host. The T-DNA first directs the plant cells to make auxins and cytokinins, which causes the cells to become irregularly shaped and form a visible tumor called a gall (Moore, L. W.). The T-DNA then directs the plant cell to start making opines (usually nopaline or agropine) which A. tumefaciens use as an energy source (Vaudequin-Dransart, V.). Since few other organisms (including the plant cell which it infects) can use the opines as an energy source, A. tumefaciens creates a special niche for itself inside the gall (Vaudequin-Dransart, V.).
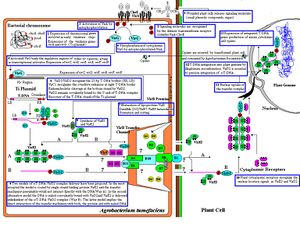
Although it is clear that A. tumefaciens integrates its own DNA into that of its hosts and induces them to make opine-producing tumor cells, the details of this process are just being unraveled. The bacteria uses a Type IV secretion system to inject the T-DNA and virulence protein into its host (Goodner, B.). It has been discovered that an entire suite of proteins, in addition to the T-DNA, is necessary to induce the host plant to make tumor cells. When the T-DNA is copied from the Ti plasmid a protein called VirD2 complexes with it to protect it from host cell nucleases and help its transport into the nucleus where it can act (Escudero, J.). In order for the T-DNA to cross from the bacteria into the plant cell, a protein called VirE2 forms a channel between the membrane of the plant and bacterial cell (Duckley, M). Another protein, VirE3, has been discovered to be injected into the plant cell. It has been shown that VirE3 is translocated into the plant nucleus where it interacts with native transcription factors to cause a deregulation of plant cell proliferation and cause the growth of galls (Garcia-Rodriguez, F. M.). Other virulence factors are not necessary for the process of tumor induction but extends the host range of A. tumefaciens. The process by which A. tumefaciens causes disease has not fully been elucidated, future research still promises to shed light on the process.
Cell structure and metabolism
A. tumefaciens contain flagella, which are important in their life cycle as it helps them swim through the soil to find their plant hosts. Mutations in flagella genes reduced virulence of A. tumefaciens in the laboratory. Strains with fla protein mutants could not infect plants grown naturally in soil (Chesnokova, O.). The bacterium swims towards concentration of phenolic compounds and also small metabolites such as glucose and amino acids which are usually exuded from plant wounds. VirA is the receptor which senses the presence of these substances and guides the flagella towards the source of these substances. Activated VirA then phosphorylates VirG, which is responsible for preparing the cell for infecting host plant tissue, including the chemotactic response of swimming towards up the gradient of phenolics to find its host (Jin, S.).
Like many other bacteria A. tumefaciens makes pili which scientists have called the T-pilus (Bulgakov, V.). In other bacteria these structures are used to transfer genetic material between bacteria during conjugation, but this bacteria adapts it to transferring genetic material to its host.
A. tumefaciens can use a variety of substrates for energy and carbon, but it is especially evolved to use a class of chemicals called opines, which are amino acid-like compounds that are intermediates of metabolism in most organisms. A. tumefaciens forces the plants it infects to produce opines, a molecule the bacteria use as an energy and carbon source (Moore, L. W.). There are many types of opines which it can use, such as nopaline, agropine, mannopine (which are common), and chrysopine, deoxy-fructosyl-oxo-proline (which are uncommon) (Moore, L. W.). It is believed that each strain of A. tumefaciens can only metabolize one type of opine, and contains genes for its synthesis (usually in its T-DNA which it transfers to its plant host) and catabolism, although this is not strictly true (Moore, L. W.).
Ecology
A. tumefaciens can live either freely in the soil or inside plants as a parasite. When it is a parasite it uses its plant host to produce energy for it. The form of energy the plant provides to the bacteria are opines, which few bacteria except for A. tumefaciens can use as their sole energy source. This gives rise to the "Opine Hypothesis" which states that A. tumefaciens is the main beneficiary of the parasitic relationship. However, the natural situation might be more complex than what the "Opine Hypothesis" suggests, since various species of Pseudomonas which also have the capability of using opines as an energy source have been discovered in opine-producing galls along with A. tumefaciens (Moore, L. W.), so the opine niche which the bacteria creates also supports the growth of other organisms.
A. tumefaciens also competes with other bacteria in the soil, including closely related bacteria. Agrobacterium radiobacter strain K84, a non-pathogenic species of the same genus, also has an unusual ability to use opines as an energy source. Being able to grow on the same opine-producing galls as A. tumefaciens, it secretes a substance called Agrocin 84, which is taken up by A. tumefaciens. Agrocin 84, a nucleotide analogue, blocks DNA replication in A. tumefaciens and eventually kills it. A. radiobacter is immune from Agrocin 84 since it contains natural pathways for detoxifying the substance. The ability for A. radiobacter to kill A. tumefaciens have been exploited by horticulturalists to control the damage done by A. tumefaciens(Ryder, M).
Application to Biotechnology
Due to its ability to integrate DNA into its plant hosts, A. tumefaciens have been used to make transgenic plants since the 1970's. Even though other methods such as biolistic has developed to put genes inside plants, A. tumefaciens has remained popular for genetic manipulation of plants due to the low copy number of genes in plants created and stability of the transgene (Li, W.). The trick is that A. tumefaciens will inject whatever DNA is flanked by a specific 25 bp border repeat sequence (Li, W.). In normal A. tumefaciens this DNA is the T-DNA, so the first step is to create an artificial plasmid with the T-DNA excised, then insert the desired gene construct with the flanking border repeats into the plasmid. The gene construct will usually contain a selection marker (Li, W.) (usually kanamycin resistance) along with the desired gene(s) to be expressed in the plant. This artificial plasmid is then transformed into A. tumefaciens, presumably one that lacks a normal Ti plasmid. Plant tissue is then infected with this engineered A. tumefaciens, placed on a selection media (such as one containing kanamycin), and using plant tissue culture methods, the selection media will have killed the cells that have not been transformed, and the shoots which have successfully grown will contain the desired gene which was inserted.
Modern methods for creating transgenic plants using A. tumefaciens is usually a variation of the method described above. In order to simplify the process of plasmid construction, a binary vector is sometimes used (Li, W.). A binary vector is simply a strain of A. tumefaciens with a Ti plasmid that contains all the genes necessary to inject DNA into the plant but contains no T-DNA with border repeats (Li, W.). Another plasmid containing the gene that is to be inserted into the plant is then put in. Since the T-DNA does not have to be on the Ti plasmid in order for it to be integrated into the host genome, it can be put on a separate plasmid as long as it as the flanking 25-bp border repeats. A previous limitation of using A. tumefaciens to create transgenic plants is that A. tumefaciens only infects dicots and gymnosperms in nature (Li, W.). However, additional methods have been discovered which extend A. tumefaciens' host range to monocots, so this method is now useful for creating transgenic plants for all flowering plants.
Current Research
A. agrobacterium continues to be one of the oldest and most commonly used technique for transfecting genes into plants. Recent research to produce cellulase has used to express a bacterial cellulase gene in Lemna minor (duckweed) using A. agrobacterium. As the interest in producing cellulosic ethanol has increase, a method for breaking down cellulose into sugars is being seeked by scientists. A gene for cellulase was isolated from a thermophilic bacteria and transformed into duckweed using A. tumefaciens as the vector. Duckweed expressing this bacterial cellulase is in no danger of destroying its cell wall since the bacterial cellulase in question is only active at 60 - 80 degrees Celsius (Sun, Y.). Because Duckweed is a plant with high protein content (up to 40% of its dry weight) it is expected that it could be turned into an efficient bioreactor for proteins such as cellulase. However, extraction of cellulase from genetically engineered duckweed has been complicated by the fact that a subunit of the cellulase complexes with the cellulose of the duckweed in a form that is hard to remove (Sun, Y.). Further engineering of the cellulase to rid the subunit which binds to cellulose will hopefully result in a higher yield of cellulase (Sun, Y.).
Even though A. tumefaciens infects only plants in nature, it has also been shown to infect other species, including fungus and even animals. Techniques have been used to knock genes into species as diverse as fungus and animals. In fact, it is believed that A. tumefaciens is capable of putting genes into most eukaryotic species, and this could be turned into a versatile genetic manipulation technique, taking advantage of the bacteria's ability to integrate only one copy of the desired gene stably into its host. When sea urchin embryos are incubated with A. tumefaciens, it was discovered that T-pili, which the bacteria uses to transfer DNA into its plant host, formed between the bacteria and the sea urchin cells (Bulgakov, V.). When the embryo spontaneously dissociated into single cells they were examined and discovered to contain T-DNA integrated into their genome. A. tumefaciens transformation could potentially complement current methods of inserting genes into eukaryotic organisms (Bulgakov, V.).
Of great interest to scientists is the molecular machinery which A. tumefaciens uses to inject DNA into its host, which has only been characterized in the last decade or so. One protein of interest is VirE2. VirE2 appears to bind to single-stranded T-DNA and appears to protect it from the external environment, but in recent years it was discovered it had another function. It appears to form a voltage-gated channel in the plant cell's plasma membrane through which single-stranded T-DNA could pass through into the plant (Duckley, M.). VirE2 therefore has two different forms with two different functions, first in its monomeric form it is freely soluble and could bind to and protect single-stranded T-DNA, while in its multimeric form it is membrane-bound and acts as a channel for which ssDNA and other soluble components could pass through (Duckley, M).
References
Edited by Allan Wu of Rachel Larsen
Edited KMG