Anabaena variabilis: Difference between revisions
No edit summary |
No edit summary |
||
| (27 intermediate revisions by 4 users not shown) | |||
| Line 1: | Line 1: | ||
{{Curated}} | |||
{{Biorealm Genus}} | {{Biorealm Genus}} | ||
| Line 4: | Line 5: | ||
==Classification== | ==Classification== | ||
===Higher order taxa=== | ===Higher order taxa=== | ||
Bacteria; Cyanobacteria; Nostocales; Nostocaceae; | Bacteria; Cyanobacteria; Nostocales; Nostocaceae; | ||
===Genus and Species=== | ===Genus and Species=== | ||
| Line 18: | Line 18: | ||
'''NCBI: [http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?mode=Tree&id=2&lvl=3&lin=f&keep=1&srchmode=1&unlock Taxonomy]''' | '''NCBI: [http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?mode=Tree&id=2&lvl=3&lin=f&keep=1&srchmode=1&unlock Taxonomy]''' | ||
|} | |} | ||
==Description and significance== | ==Description and significance== | ||
''Anabaena variabilis'' is a bacteria strain of blue-green algae that are known as cyanobacteria. Cyanobacteria are able to carry out photosynthesis, fix nitrogen from the environment, and also produce hydrogen. This feature of creating hydrogen with energy from the sun has made it popular in many laboratories in the world. The main role in its sequencing and research is because of the previous data collected regarding its physiology, biochemistry, and genetics can be better understood (4). This particular strain has also been described to have a “complex life cycle” that involves many types of differentiated cells. The cells described were the “Heterocysts for nitrogen fixation, akinetes (spores) for survival, and hormogonia for motility and establishment of symbiotic associations with plants and fungi” (4). <br /> | |||
<br />The organism ''Anabaena variabilis'' was isolated at the U.S. Department of Energy Joint Genome Institute (DOE JGI). The DOE JGI listed that it sequenced the genomes of the organisms through the use of “whole-genome shotgun” (5). It described the technique as randomly “shearing” the genome DNA and then re-assembling it by comparing similar sequences and then inferring the original genome. The institute lists the three different size fragments that it can use and gives an example of how the procedure is done (5). | |||
==Genome structure== | ==Genome structure== | ||
''Anabaena variabilis'' has been sequenced and the data gathered show that the circular DNA contains 6,365,727 base pairs (2). The information on the site shows that the GC-content of the organism is at 41.4% and that the genome as of now codes for 5043 proteins and 62 RNA strands (3). The cyanobacteria also have three plasmids which are named plasmid A, B, and C (3). The plasmids probably affect the organism’s ability to produce immunity to harmful substances and also provide the codes for certain proteins. Respectively, the plasmids each code for 344, 31 and 243 proteins within the cyanobacteria (3). <br /> <br /> | |||
Some interesting characteristics about this organism are some of its genes. It contains two gene clusters, ''nif1'' and ''nif2'', that code for Mo-nitrogenase (7). It also has another gene cluster, ''vnf'', which codes for V-Fe cofactor nitrogenase. Each of the nitrogenases are activated under different conditions allowing the cyanobacteria to function. The ''Anabaena variabilis'' is also one of the few prokaryotic organisms that contains the enzyme phenylalanine ammonia lyase (PAL), which helps deaminate phenylalanine to cinnamate (6). | |||
==Cell structure and metabolism== | ==Cell structure and metabolism== | ||
As described before, the interesting features of ''Anabaena variabilis'' stem from its genes, which code for proteins that affect the whole organism. The different nitrogenase enzymes that are produced allow the organism to live in different environments, while still carrying out its major processes. The different conditions for each gene are specified below: <br /> <br /> | |||
''nif1'': “diazotrophic conditions only in heterocysts” <br /> | |||
''nif2'': “strictly anoxic conditions in vegetative and heterocysts” <br /> | |||
''vnf'': “diazotrophic conditions lacking molybdate, with or without vanadate” (7). <br /> <br /> | |||
Under the first gene, the organism can grow without having the need of a fixed nitrogen source. The organism is able to fix its own nitrogen from the environment by using the Mo-Fe cofactor (7). | |||
With the second gene the cell can continue to grow even without an oxygen source. The organism will still use the same Mo-Fe cofactor. The third gene that is transcribed to fix nitrogen will occur when the organism is exposed to an environment lacking molybdate, which is part of the Mo-Fe cofactor needed with the genes ''nif1'' and ''nif2''. This allows the organism to grow without the use of a certain cofactor that other cyanobacteria would need. <br /> <br /> | |||
Another interesting feature about the ''Anabaena variabilis'' is that it has the gene that codes for the production of the phenylalanine ammonia lyase (PAL). This enzyme is common in terrestrial plants, but rare in prokaryotic organisms. It is used to catalyze the beginning of the formation of phenylpropanoids (6). | |||
==Ecology== | ==Ecology== | ||
Cyanobacteria are described as “blue-green algae” and therefore found in an aquatic based environment such as the oceans or other bodies of water. It is believed that cyanobacteria on Earth are responsible as being the producer for most, if not all, the oxygen in the atmosphere (8). Plants also produce oxygen, however, they depend on chloroplasts. These plant organelles are believed to have been derived from the cyanobacteria (8). The cyanobacteria seem to have been the foundation of changing the Earth’s atmosphere because it takes care of half of the Earth’s photosynthesis (8). | |||
==Pathology== | ==Pathology== | ||
''Anabaena variabilis'' is not classified as a pathogen. However, the cyanobacteria is known to produce “toxic blooms” in their environment. The products are dangerous or deadly to animals and humans due to various cyanotoxins that are released (3). | |||
==Application to Biotechnology== | ==Application to Biotechnology== | ||
The ''Anabaena variabilis'' strain has been extensively studied because it also undergoes a process where it produces hydrogen gas by using sunlight (4). This product could provide a reusable source of energy. By further studying the genome and understanding the process, the hydrogen gas could be mass produced and used as fuel or energy. <br /> <br /> | |||
A more recent study indicated that the Phenylalanine ammonia lyase (PAL), could be a potential solution to the disease known as phenylketonuria. Current research is being done on the possible effects on animals and how they would carry over to humans (6). | |||
==Current Research== | ==Current Research== | ||
The research concerning ''Anabaena variabilis'' and how it’s one of the few prokaryotic organisms that produce the enzyme phenylalanine ammonia lyase (PAL) indicates how it could be used to treat a metabolic disease called phenylketonuria (PKU). The disorder causes a buildup of phenylalanine due to a mutation in an enzyme that would normally break down the phenylalanine. This leads up to a dangerous build up of neurotoxins that can lead to serious health hazards or even death. The studies so far have shown that PAL will reduce the amount phenylalanine in the blood. However, the tests also show that there are problems regarding stability of the enzyme, its administration, and immunological response to the new enzyme (6). <br /> <br /> | |||
Recent studies have been using 2-dimensional electrophoresis (2-DE) in order to further understand some of the proteins within the organism. A 2-DE study will separate the proteins according to two different properties with each separation being perpendicular to the other. For example, if one property separated the molecules horizontally, the second property would separate the molecules vertically. This study resulted in the identification of 254 unique proteins. Further comparisons showed other properties of the ''Anabaena variabilis'' (1). <br /> <br /> | |||
Another study has been focused on identifying factors affecting the growth of ''Anabaena variabilis'' in a bubble column reactor. The factors that focused on were the “superficial gas velocity, pH, initial phosphate concentration, and light intensity” in order to assess the mass production of cyanobacteria. The results showed that the best conditions for growth was when superficial gas velocity was at 2.0cm/s, pH 7.0, initial phosphate concentration of 55mg/L, with a irradiation rate (how they controlled light intensity) above 11.5mumol/s/g (9). | |||
==References== | |||
1. [http://www3.interscience.wiley.com/cgi-bin/fulltext/114212206/PDFSTART Barrios-Llerena, M. E., Reardon, K. F., and Wright, P. C. “2-DE proteomic analysis of the model cyanobacterium ''Anabaena variabilis''”. Electrophoresis. 2007. Vol. 28. p. 1624-1632.] | |||
<br /> <br /> | |||
2. [http://www.ncbi.nlm.nih.gov/entrez/viewer.fcgi?db=nuccore&id=75699950 Genome of ''Anabaena variabilis'' (NCBI Sequence Viewer)] | |||
<br /> <br /> | |||
3. [http://www.ncbi.nlm.nih.gov/sites/entrez?db=genomeprj&cmd=Retrieve&dopt=Overview&list_uids=10642 Genome Project Results (NCBI)] | |||
<br /> <br /> | |||
4. [http://genome.jgi-psf.org/finished_microbes/anava/anava.home.html JGI ''Anabaena variabilis'' ATCC 29413] | |||
<br /> <br /> | |||
5. [http://www.jgi.doe.gov/education/how/ JGI Education How] | |||
<br /> <br /> | |||
6. [http://pubs.acs.org/cgi-bin/article.cgi/bichaw/2007/46/i04/html/bi061774g.html Moffitt, M. C., Louie, G. V., Bowman, M. E., Pence, J. Noel, J. P. and Moore, B. S. “Discovery of Two Cyanobacterial Phenylalanine Ammonia Lyases: Kinetic and Structural Characterization”. Biochemistry. 2007. Vol. 46. p. 1004-1012.] | |||
<br /> <br /> | |||
7. [http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pubmed&pubmedid=16885448 Pratte, B. S., Eplin, K., and Thiel, T. “Cross-Functionality of Nitrogenase Components NifH1 and VnfH in ''Anabaena variabilis''”. Journal of Bacteriology. 2006. Vol. 188, No. 16. p.5806-5811.] | |||
<br /> <br /> | |||
8. Schaechter, M., Ingraham, J. L., Neidhardt, F. C. “Prokaryotic Microbes: Phylum B10: Cyanobacteria”. Microbe. 2006. Ch. 15. p. 306-307. | |||
<br /> <br /> | |||
9. [http://www.sciencedirect.com/science?_ob=ArticleURL&_udi=B6V24-4NBRFPB-5&_user=4429&_coverDate=03%2F26%2F2007&_rdoc=1&_fmt=&_orig=search&_sort=d&view=c&_acct=C000059602&_version=1&_urlVersion=0&_userid=4429&md5=7ba5735c108ea7b35818fda19185b77c Yoon, J. H., Shin, J.-H., and Park, T. H. “Characterization of factors influencing the growth of ''Anabaena variabilis'' in a bubble column reactor”. Bioresource Technology. 2007.] | |||
<br /> <br /> | |||
10. '''Copyright Status''' <br /> | |||
Joint Genome Institute authored documents are sponsored by the U.S. Department of Energy under Contracts W-7405-Eng-48, DE-AC02-05CH11231, and W-7405-ENG-36. Accordingly, the U.S. Government retains a nonexclusive, royalty-free license to publish or reproduce these documents, or allow others to do so, for U.S. Government purposes. All documents available from this server may be protected under the U.S. and Foreign Copyright Laws and permission to reproduce them may be required. The public may copy and use this information without charge, provided that this Notice and any statement of authorship are reproduced on all copies. JGI is not responsible for the contents of any off-site pages referenced. | |||
<br /> | |||
Edited by Phuoc Nam Nguyen, student of [mailto:ralarsen@ucsd.edu Rachel Larsen] and Kit Pogliano | Edited by Phuoc Nam Nguyen, student of [mailto:ralarsen@ucsd.edu Rachel Larsen] and Kit Pogliano | ||
Latest revision as of 15:14, 7 July 2011
A Microbial Biorealm page on the genus Anabaena variabilis
Classification
Higher order taxa
Bacteria; Cyanobacteria; Nostocales; Nostocaceae;
Genus and Species
Anabaena variabilis
|
NCBI: Taxonomy |
Description and significance
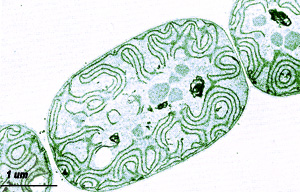
Anabaena variabilis is a bacteria strain of blue-green algae that are known as cyanobacteria. Cyanobacteria are able to carry out photosynthesis, fix nitrogen from the environment, and also produce hydrogen. This feature of creating hydrogen with energy from the sun has made it popular in many laboratories in the world. The main role in its sequencing and research is because of the previous data collected regarding its physiology, biochemistry, and genetics can be better understood (4). This particular strain has also been described to have a “complex life cycle” that involves many types of differentiated cells. The cells described were the “Heterocysts for nitrogen fixation, akinetes (spores) for survival, and hormogonia for motility and establishment of symbiotic associations with plants and fungi” (4).
The organism Anabaena variabilis was isolated at the U.S. Department of Energy Joint Genome Institute (DOE JGI). The DOE JGI listed that it sequenced the genomes of the organisms through the use of “whole-genome shotgun” (5). It described the technique as randomly “shearing” the genome DNA and then re-assembling it by comparing similar sequences and then inferring the original genome. The institute lists the three different size fragments that it can use and gives an example of how the procedure is done (5).
Genome structure
Anabaena variabilis has been sequenced and the data gathered show that the circular DNA contains 6,365,727 base pairs (2). The information on the site shows that the GC-content of the organism is at 41.4% and that the genome as of now codes for 5043 proteins and 62 RNA strands (3). The cyanobacteria also have three plasmids which are named plasmid A, B, and C (3). The plasmids probably affect the organism’s ability to produce immunity to harmful substances and also provide the codes for certain proteins. Respectively, the plasmids each code for 344, 31 and 243 proteins within the cyanobacteria (3).
Some interesting characteristics about this organism are some of its genes. It contains two gene clusters, nif1 and nif2, that code for Mo-nitrogenase (7). It also has another gene cluster, vnf, which codes for V-Fe cofactor nitrogenase. Each of the nitrogenases are activated under different conditions allowing the cyanobacteria to function. The Anabaena variabilis is also one of the few prokaryotic organisms that contains the enzyme phenylalanine ammonia lyase (PAL), which helps deaminate phenylalanine to cinnamate (6).
Cell structure and metabolism
As described before, the interesting features of Anabaena variabilis stem from its genes, which code for proteins that affect the whole organism. The different nitrogenase enzymes that are produced allow the organism to live in different environments, while still carrying out its major processes. The different conditions for each gene are specified below:
nif1: “diazotrophic conditions only in heterocysts”
nif2: “strictly anoxic conditions in vegetative and heterocysts”
vnf: “diazotrophic conditions lacking molybdate, with or without vanadate” (7).
Under the first gene, the organism can grow without having the need of a fixed nitrogen source. The organism is able to fix its own nitrogen from the environment by using the Mo-Fe cofactor (7).
With the second gene the cell can continue to grow even without an oxygen source. The organism will still use the same Mo-Fe cofactor. The third gene that is transcribed to fix nitrogen will occur when the organism is exposed to an environment lacking molybdate, which is part of the Mo-Fe cofactor needed with the genes nif1 and nif2. This allows the organism to grow without the use of a certain cofactor that other cyanobacteria would need.
Another interesting feature about the Anabaena variabilis is that it has the gene that codes for the production of the phenylalanine ammonia lyase (PAL). This enzyme is common in terrestrial plants, but rare in prokaryotic organisms. It is used to catalyze the beginning of the formation of phenylpropanoids (6).
Ecology
Cyanobacteria are described as “blue-green algae” and therefore found in an aquatic based environment such as the oceans or other bodies of water. It is believed that cyanobacteria on Earth are responsible as being the producer for most, if not all, the oxygen in the atmosphere (8). Plants also produce oxygen, however, they depend on chloroplasts. These plant organelles are believed to have been derived from the cyanobacteria (8). The cyanobacteria seem to have been the foundation of changing the Earth’s atmosphere because it takes care of half of the Earth’s photosynthesis (8).
Pathology
Anabaena variabilis is not classified as a pathogen. However, the cyanobacteria is known to produce “toxic blooms” in their environment. The products are dangerous or deadly to animals and humans due to various cyanotoxins that are released (3).
Application to Biotechnology
The Anabaena variabilis strain has been extensively studied because it also undergoes a process where it produces hydrogen gas by using sunlight (4). This product could provide a reusable source of energy. By further studying the genome and understanding the process, the hydrogen gas could be mass produced and used as fuel or energy.
A more recent study indicated that the Phenylalanine ammonia lyase (PAL), could be a potential solution to the disease known as phenylketonuria. Current research is being done on the possible effects on animals and how they would carry over to humans (6).
Current Research
The research concerning Anabaena variabilis and how it’s one of the few prokaryotic organisms that produce the enzyme phenylalanine ammonia lyase (PAL) indicates how it could be used to treat a metabolic disease called phenylketonuria (PKU). The disorder causes a buildup of phenylalanine due to a mutation in an enzyme that would normally break down the phenylalanine. This leads up to a dangerous build up of neurotoxins that can lead to serious health hazards or even death. The studies so far have shown that PAL will reduce the amount phenylalanine in the blood. However, the tests also show that there are problems regarding stability of the enzyme, its administration, and immunological response to the new enzyme (6).
Recent studies have been using 2-dimensional electrophoresis (2-DE) in order to further understand some of the proteins within the organism. A 2-DE study will separate the proteins according to two different properties with each separation being perpendicular to the other. For example, if one property separated the molecules horizontally, the second property would separate the molecules vertically. This study resulted in the identification of 254 unique proteins. Further comparisons showed other properties of the Anabaena variabilis (1).
Another study has been focused on identifying factors affecting the growth of Anabaena variabilis in a bubble column reactor. The factors that focused on were the “superficial gas velocity, pH, initial phosphate concentration, and light intensity” in order to assess the mass production of cyanobacteria. The results showed that the best conditions for growth was when superficial gas velocity was at 2.0cm/s, pH 7.0, initial phosphate concentration of 55mg/L, with a irradiation rate (how they controlled light intensity) above 11.5mumol/s/g (9).
References
1. Barrios-Llerena, M. E., Reardon, K. F., and Wright, P. C. “2-DE proteomic analysis of the model cyanobacterium Anabaena variabilis”. Electrophoresis. 2007. Vol. 28. p. 1624-1632.
2. Genome of Anabaena variabilis (NCBI Sequence Viewer)
3. Genome Project Results (NCBI)
4. JGI Anabaena variabilis ATCC 29413
5. JGI Education How
6. Moffitt, M. C., Louie, G. V., Bowman, M. E., Pence, J. Noel, J. P. and Moore, B. S. “Discovery of Two Cyanobacterial Phenylalanine Ammonia Lyases: Kinetic and Structural Characterization”. Biochemistry. 2007. Vol. 46. p. 1004-1012.
7. Pratte, B. S., Eplin, K., and Thiel, T. “Cross-Functionality of Nitrogenase Components NifH1 and VnfH in Anabaena variabilis”. Journal of Bacteriology. 2006. Vol. 188, No. 16. p.5806-5811.
8. Schaechter, M., Ingraham, J. L., Neidhardt, F. C. “Prokaryotic Microbes: Phylum B10: Cyanobacteria”. Microbe. 2006. Ch. 15. p. 306-307.
9. Yoon, J. H., Shin, J.-H., and Park, T. H. “Characterization of factors influencing the growth of Anabaena variabilis in a bubble column reactor”. Bioresource Technology. 2007.
10. Copyright Status
Joint Genome Institute authored documents are sponsored by the U.S. Department of Energy under Contracts W-7405-Eng-48, DE-AC02-05CH11231, and W-7405-ENG-36. Accordingly, the U.S. Government retains a nonexclusive, royalty-free license to publish or reproduce these documents, or allow others to do so, for U.S. Government purposes. All documents available from this server may be protected under the U.S. and Foreign Copyright Laws and permission to reproduce them may be required. The public may copy and use this information without charge, provided that this Notice and any statement of authorship are reproduced on all copies. JGI is not responsible for the contents of any off-site pages referenced.
Edited by Phuoc Nam Nguyen, student of Rachel Larsen and Kit Pogliano
