Aspergillus tubingensis
1. Classification
Higher order taxa
Eukarya; Fungi; Dikarya; Ascomycota; Pezizomycotina; Eurotiomycetes; Eurotiomycetidae; Eurotiales; Aspergillaceae; Aspergillus; Tubingensis
Species
|
NCBI: [1] |
Aspergillus tubingensis
2. Description and significance
Description
Aspergillus tubingensis is a darkly pigmented fungus with a rough texture [2]. As an asexual reproductive fungus, it produces spores that start germinating upon release and generate hyphae that make adult mycelium [2],[3]. This fungus is often associated with Aspergillus niger since they have similar morphologies [2]. Molecular tools such as PCR analysis are helpful in distinguishing the two taxa successfully [2],[4].
Significance
Aspergillus tubingensis is often associated with the spoilage of food products [5]. It has also been shown to be an opportunistic pathogen of humans and can cause skin infections and otomycosis [6],[7]. A. tubingensis is significant due to its ability to degrade polyester polyurethane plastics and has been extensively studied for its potential to reduce the contamination of water and soil [8]. It has the potential to reduce human waste and associated public health hazards linked with plastic toxicity [8],[9]. A. tubingensis is also significant in the agricultural field due to its ability to protect plants from fungal diseases by producing glucose oxidases and other metabolites that can be used as biofungicides [10. Commercially, A. tubingensis is being investigated as a replacement for chemical products to fight tropical bed bugs [11].
3. Genome structure
The genome sizes of Aspergillus tubingensis strains G131 and CBS 134.48 are 35,18 Mb and 35,15 Mb respectively. Strain G131 consists of 10,994 coding genes with an average protein length of 522 amino acids [12]. Strain CBS 134.48 consists of 12,322 coding genes with an average protein length of 465 amino acids [12]. The GC content in both strains is 50.22% and 49.18% respectively [12]. Both strains show gene clusters with useful functions in the synthesis of cytochrome P450, carboxylesterases, methyltransferases, and other secondary metabolites [12]. The genome of strain G131 consists of 80 secondary metabolite gene clusters. Comparing its genome with other black Aspergilli - A. tubingensis CBS 134.48, A. niger, and A. kawachii, shows 7 unique clusters in addition to some conserved clusters [12]. Genes coding for ochratoxin A (OTA) and fumonisins, mycotoxins synthesized by other black Aspergilli, are absent in A. tubingensis G131 [12]. However, genome analysis of this strain indicates its potential in the synthesis of metabolites for the production of antibiotics and antioxidants [12].
4. Cell structure
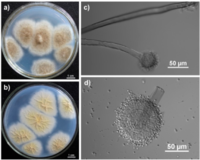
Aspergillus tubingensis is a hyphal fungus that forms black colonies when grown on CYA (Czapeck Yeast Agar) culture medium [13]. It produces spores in the form of conidia that range from 4-5 µm in diameter with a wrinkled texture, globular shape, and vesicles that are 45-69 µm in size [13. The morphology of A. tubingensis closely resembles that of A. niger. The formation of whitish-pink sclerotia by most A. tubingensis strains distinguishes it from other morphologically similar fungi that form black colonies, including Aspergillus niger. [12],[13].
5. Metabolic processes
Aspergillus tubingensis metabolizes carbohydrates, lipids, and amino acids, in addition to other secondary metabolites [12] . It usually grows on carbon-rich substrates such as glucose and amylose. With creatine sucrose agar (CREA) culture medium, A. tubingensis demonstrates good acid production and a moderate growth rate [12]. Due to its potential to carry out secondary metabolism, A. tubingensis is important for the production of secondary metabolites in the industry of biotechnology [12]. A. tubingensis G131 can synthesize metabolites such as TAN-1612, naphtha-gamma-pyrones, and asperazine for the production of antibiotics, antioxidants, and anticancer molecules [12]. Frequent food contaminants such as ochratoxin A (OTA) and fumonisins are absent in A. tubingensis [12].
6. Ecology
Aspergillus tubingensis can survive dry conditions and high temperatures above 45 °C [16]. It typically lives in the soil and in crops such as maize and wheat [17]. This fungus lives within plants and has been found in Pongamia pinnata of mangroves [18]. It is also present in certain aquatic organisms such as the crab Portunus trituberculatus [19]. This fungus can be found as contaminants in dry-fruits such as raisins [5]. It can absorb heavy metal toxins that are present in soils [20]. A. tubingensis also acts as an additive to foods, such as breads, when paired with ascorbic acid and amylase [15]. However, it can also infect humans [6]. For example, Aspergillus tubingensis can live in the lungs, resulting in an infection, especially for those suffering from Cystic Fibrosis [21].
7. Pathogenicity
Aspergillus tubingensis has recently been found to cause opportunistic infections. It can reside in major airways in the lungs, causing bronchial colonization in patients with pulmonary conditions [21]. It was first identified in an infection of the jaw, although it is unclear whether A. tubingensis was responsible for the initial infection [6]. In another study, A. tubingensis was found to be the cause of an opportunistic skin infection in a diabetic patient [7]. It was identified as a member of the Aspergillus nigri section by size, temperature tolerance, response to antifungals, and its appearance as black colonies surrounded by white mycelium [7]. A. tubingensis responds to the antifungals typically used to treat Aspergillus nigri infections [7].
8. Current Research
Aspergillus tubingensis is being studied for its ability to decompose plastics [9],[13]. Although it has been found to biodegrade polyester polyurethane and high density polyethylene, these studies have been small-scale and research is still being conducted to test its commercial viability [9]. Aspergillus tubingensis is also being investigated as a commercially feasible treatment for effective wastewater management, improved production of biofuels, and reduced plastic pollution. For example, A. tubingensis can potentially be used to speed up the production of aerobic granular sludge, which is used in wastewater treatment to remove pollutants from sewage [3],[22]. A. tubingensis can also biodegrade sugarcane bagasse into bio-ethanol [6],[23].
9. References
[1] National Center for Biotechnology Information. n. d. Taxonomy Database; txid=5068, https://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?mode=Info&id=5068&lvl=3&lin=f&keep=1&srchmode=1&unlock [Taxonomy] (Accessed Oct 15, 2020).
[2] Samson, R. A., Noonim, P., Meijer, M., Houbraken, J., Frisvad, J. C., and Varga, J. 2007. Diagnostic tools to identify black aspergilli. Studies in mycology 59: 129-145. https://doi.org/10.3114/sim.2007.59.13
[3] Aspergillus Tubingensis. n.d. [Revised June 11, 2020]. In Wikipedia the free encyclopedia. https://en.wikipedia.org/wiki/Aspergillus_tubingensis [Wiki]. (Accessed Sep 17, 2020).
[4] Palumbo, J. D., and O’Keeffe, T.L. 2015. Detection and discrimination of four Aspergillus section Nigri species by PCR. Letters in applied microbiology 60(2): 188-195. https://doi.org/10.1111/lam.12358
[5] Tournas, V. H., Niazi, N. S., and Kohn, J. S. 2015. Fungal presence in selected tree nuts and dried fruits. Microbiology insights 8: 1–6. https://doi.org/10.4137/MBI.S24308
[6] Bathoorn, E., Escobar Salazar, N., Sepehrkhouy, S., Meijer, M., de Cock, H., & Haas, P. J. 2013. Involvement of the opportunistic pathogen Aspergillus tubingensis in osteomyelitis of the maxillary bone: a case report. BMC infectious diseases 13, 59. https://doi.org/10.1186/1471-2334-13-59
[7] Frías-De-León, M. G., Rosas-de Paz, E., Arenas, R., Atoche, C., Duarte-Escalante, E., Molina de Soschin, D., Acosta-Altamirano, G., and Reyes-Montes, M. R. 2018. Identification of Aspergillus tubingensis in a primary skin infection. Journal de mycologie medicale 28(2): 274–278. https://doi.org/10.1016/j.mycmed.2018.02.013
[8] Khan, S., Nadir, S., Shah, Z. U., Shah, A. A., Karunarathna, S. C., Xu, J., Khan, A., Munir, S., and Hasan, F. 2017. Biodegradation of polyester polyurethane by Aspergillus tubingensis. Environmental pollution 225: 469–480. https://doi.org/10.1016/j.envpol.2017.03.012
[9] Sangeetha Devi, R., Rajesh Kannan, V., Nivas, D., Kannan, K., Chandru, S., and Robert Antony, A. 2015. Biodegradation of HDPE by Aspergillus spp. from marine ecosystem of Gulf of Mannar, India. Marine pollution bulletin 96(1-2): 32–40. https://doi.org/10.1016/j.marpolbul.2015.05.050
[10] Kriaa, M., Hammami, I., Sahnoun, M., Azebou, M. C., Triki, M. A., and Kammoun, R. 2015. Biocontrol of tomato plant diseases caused by Fusarium solani using a new isolated Aspergillus tubingensis CTM 507 glucose oxidase. Comptes rendus biologies 338(10): 666–677. https://doi.org/10.1016/j.crvi.2015.05.007
[11] Zahran, Z., Nor, N. M. I. Z., Dieng, H., and Satho, T. 2017. Laboratory Efficacy of Mycoparasitic Fungi (Aspergillus Tubingensis and Trichoderma Harzianum) against Tropical Bed Bugs (Cimex Hemipterus) (Hemiptera: Cimicidae). Asian Pacific Journal of Tropical Biomedicine 7(4): 288–293. http://dx.doi.org/10.1016/j.apjtb.2016.12.021
[12] Choque, E., Klopp, C., Valiere, S., Raynal, J., and Mathieu, F. 2018. Whole-genome sequencing of Aspergillus tubingensis G131 and overview of its secondary metabolism potential. BMC genomics 19(1): 200. https://doi.org/10.1186/s12864-018-4574-4
[13] Silvia, D. M., Batista, L. R., Rezende, E. F., Fungaro, M. H., Sartori, d., and Alves, E. 2007. Identification of fungi of the genus Aspergillus section nigri using polyphasic taxonomy. Brazilian journal of microbiology 49(2): 761-773. https://doi.org/10.1590/S1517-838220110002000044
[14] File: Aspergillus tubingensis FJBJ11.png. n.d. [Revised Oct 5, 2020]. In Wikimedia Commons, the free media repository. https://commons.wikimedia.org/wiki/File:Aspergillus_tubingensis_FJBJ11.png [Wiki]. (Accessed Nov 23, 2020).
[15] Kriaa, M., Ouhibi, R., Graba, H., Besbes, S., Jardak, M., and Kammoun, R. 2016. Synergistic effect of Aspergillus tubingensis CTM 507 glucose oxidase in presence of ascorbic acid and alpha amylase on dough properties, baking quality and shelf life of bread. Journal of food science and technology 53(2): 1259-1268. https://doi.org/10.1007/s13197-015-2092-9
[16] García-Cela, E., Crespo-Sempere, A., Ramos, A. J., Sanchis, V., and Marin, S. 2014. Ecophysiological characterization of Aspergillus carbonarius, Aspergillus tubingensis and Aspergillus niger isolated from grapes in Spanish vineyards. International journal of food microbiology 173: 89–98. https://doi.org/10.1016/j.ijfoodmicro.2013.12.012
[17] Singh, H.. and Reddy, S. M. 2011. Improvement of wheat and maize crops by inoculating Aspergillus spp. In alkaline soil fertilized with rock phosphate. Archives of Argonomy and Soil Science 58(5): 535-546 https://doi.org/10.1080/03650340.2010.532125
[18] Huan, H., Feng, X.J., Liu, L., Chen, B., Lu, Y.J., Ma, L., She, Z.G., and Lin, Y.C., 2010. Three Dimeric Naphtho-γ-Pyrones from the Mangrove Endophytic Fungus Aspergillus tubingensis Isolated from Pongamia pinnata. Planta Med 76: 1888–1891. http://dx.doi.org/10.1055/s-0030-1249955
[19] Guo L., Wang C., Wen-cheng Z., and Xu, F. 2016. Bioassay-guided fractionation and identification of active substances from the fungus Aspergillus tubingensis against Vibrio anguillarum. Biotechnology & Biotechnological Equipment, 30:3, 602-606. https://doi.org/10.1080/13102818.2016.1146635
[20] Tang, A., Lu, Y., Li, Q., Zhang, X., Cheng, N., Liu, H., and Liu, Y. 2020. Simultaneous leaching of multiple heavy metals from a soil column by extracellular polymeric substances of Aspergillus tubingensis F12. Chemosphere 263:127883. https://doi.org/10.1016/j.chemosphere.2020.127883
[21] Gautier, M., Normand, A. C., L'Ollivier, C., Cassagne, C., Reynaud-Gaubert, M., Dubus, J. C., Brégeon, F., Hendrickx, M., Gomez, C., Ranque, S., and Piarroux, R. 2016. Aspergillus tubingensis: a major filamentous fungus found in the airways of patients with lung disease. Medical mycology 54(5): 459–470. https://doi.org/10.1093/mmy/myv118
[22] Chen, Y., Ge, J., Wang, S., and Su, H. 2020. Insight into formation and biological characteristics of Aspergillus tubingensis-based aerobic granular sludge (AT-AGS) in wastewater treatment. The Science of the total environment 739: 140128. https://doi.org/10.1016/j.scitotenv.2020.140128
[23] Prajapati, B. P., Jana, U. K., Suryawanshi, R. K., and Kango, N. 2020. Sugarcane bagasse saccharification using Aspergillus tubingensis enzymatic cocktail for 2G bio-ethanol production. Renewable Energy 152: 653-663. https://doi.org/10.1016/j.renene.2020.01.063
Edited by Rashi Purohit Yara Manna, students of Jennifer Bhatnagar for BI 311 General Microbiology, 2020, Boston University.
Author contributions: R.P wrote the Classification section; Y.M wrote the Introduction section; R.P wrote the sections on Genome Structure and Cell Structure; D.L. wrote the section on Metabolic Processes; N.P wrote the sections on Pathology and Ecology; D.A. and D.L. wrote the sections on Application and Current Research; R.P. and D.A. curated figures; R.P wrote the References section; R.P. and D.A. edited final article draft.
