Bacterial Transcription Factors against Reactive Oxygen Species
Overview
By Jiayu Chen
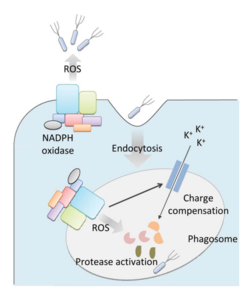
Bacteria live in a world full of toxic reactive oxygen species (ROS), including hydrogen peroxide (H2O2), superoxide (O2-), and hydroxyl radicals. ROS can cause severe damage to all kinds of cell components, including nucleic acids, lipids and proteins [1]. These highly reactive species are generated endogenously by accidental autoxidation of flavoproteins, a group of non-respiratory redox enzymes. [2] Thus, aerobic bacteria have evolved to maintain a high concentration of enzymes that keep intracellular ROS below the lethal level. However, various conditions are known to increase these species above this threshold, inflicting oxidative stress to the bacteria through a phenomenon called “oxidative burst.” [3] One of the major ways in which the bacteria encounter ROS is when ROS are generated by the immune system of their host as a defensive weapon. In humans, phagocytes including neutrophils use NADPH oxidase 2 (NOX2) complex as one of the major enzymes mediating the immune response by generating antimicrobial ROS. The resultant ROS is secreted either outside of the cell as a trap to kill the bacteria, or into phagolysosomes where internalized bacteria would be degraded (Figure 1). Another situation that may happen during bacterial infection is ROS generated by the mitochondria. This defensive mechanism is mediated by many signaling molecules in the Toll pathways including several Toll-like receptors (TLRs), tumor necrosis factor receptor-associated factor 6 (TRAF6) and the evolutionarily conserved signaling intermediates in Toll pathways (ECSIT).
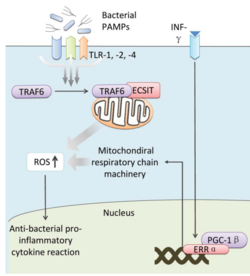
The activation of Toll pathways results in increased cellular ROS as well as anti-bacterial pro-inflammatory cytokine reactions in the nucleus. Pro-inflammatory Th1 cytokine IFN-γ is also involved as a transcriptional regulator via activation of estrogen-related receptor α (ERRα) and PPARγ-coactivator-1β (PGC-1β) in the nucleus. Such interaction induces the genes that encode mitochondrial respiratory chain machinery and thus increase cellular ROS concentration (Figure 2). [4]
Bacteria have developed defensive systems to deal with such situations. One mechanism is the ROS-inducible transcriptional factors that regulate gene expressions of various enzymes to respond to oxidative stress. Three redox-sensitive transcription factors in bacteria have been intensively studied: OxyR and PerR against H2O2, and SoxR against O2-. [5]
OxyR is induced by H2O2 and activated by reversible disulfide bond formation in its active site. [6] OxyR is widely distributed in Gram-negative bacteria, such as E. coli. The binding of OxyR to DNA generally turns on the gene expression of a large range of oxidative stress defense enzymes. [7] Recent research also has shown that Gram-negative bacteria Vibrio vulnificus, facultative aerobes, contain two OxyR-type regulators using similar sensory mechanisms but with different sensitivity in order to fine-tune the expression of genes against H2O2 pressure. [8]
PerR is the equivalent H2O2 sensor protein in many Gram-positive bacteria, such as Staphylococcus aureus and Bacillus subtilis. When the PerR protein is in its reduced form, its binding to DNA generally serves as an inhibitor of the expression of target genes. PerR is activated by oxidation of its regulatory metal, either Fe2+ or Mn2+, which leads to conformational change in the protein and dissociation from DNA, thus relieving the inhibition and initiating transcription of H2O2 defensive genes. [9] [10]
SoxR is conserved widely in both proteobacteria and actinobacteria with an O2- sensitive metal complex in its active site. In E. coli, the binding of SoxR to DNA induces the transcription of a secondary transcription factor, SoxS, which activates a group of enzymes suppressing the level of O2-. [11] [12] [13]
Defending against hydrogen peroxide (H2O2)
Why is H2O2 poisonous?
Environmental H2O2 is primarily generated as a by-product of metabolism and as a defense mechanism of plants, animals, and bacteria against invasive microbes. [14] [15] [16] Because its chemical structure is similar to that of water and has no charge, H2O2 can easily penetrate the membrane. Once it enters the cell, H2O2 damages the cell components through the Fenton Reaction in which H2O2 oxidizes Fe2+ ion, generating one Fe3+ ion and one hydroxyl radical. Free hydroxyl radicals can further react with lipids, nucleic acids and disulfide bonds in proteins, which are all critical cellular components. [17] For example, guanine, one of the four main nucleobases, is one of the most vulnerable targets to hydroxyl radicals because of its small reduction potential. The resulting product is often highly mutagenic 8-hydroxyguanine because it is able to pair with adenine in such a way that allows it to escape from DNA mismatch detection systems. [18] The Fenton reaction also impedes the function of Fe2+-dependent enzymes that are distributed in various cellular processes. [19]
Defensive strategy for Gram-negative bacteria: OxyR
H2O2 sensory mechanism of OxyR
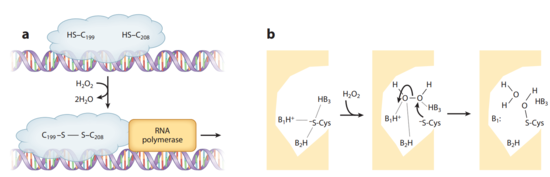
The crystal structures of OxyR in different bacteria have shown that it is a tetrameric protein. Each monomer contains a C-terminal regulatory domain and an N-terminal DNA-binding domain. The redox-switch mechanism in OxyR depends on two H2O2 sensory cysteine residues (Cys-199 and Cys-208) in its C-terminal. Cys-199 residue locates in a hydrophobic pocket, where it is stabilized by hydrophobic interactions with nearby leucine residues. Cys-199 is rapidly oxidized by H2O2 and forms a sulfenic acid intermediate (-SOH). This intermediate is not stable in the hydrophobic pocket, and thus leads to a conformational change that flips out the oxidized Cys-199. A flexible amino acid region between these two cysteine residues then brings Cys-199 to the proximity of Cys-208 to form a disulfide bond. This bond formation induces a conformational change in OxyR regulatory domain, recruiting an RNA polymerase and thus stimulating the expression of genes against H2O2 stress (Figure 3). [20] [21] Furthermore, the interaction between the OxyR and the RNA polymerase seems to be able to reduce the disulfide bond and makes the transcriptional regulation reversible. [22]
OxyR in E. coli
As a model organism in microbiology study, the OxyR of E. coli has been extensively studied in the past decades. A genome-wide transcription profile of E. coli using DNA microarray has shown that 23 genes are significantly induced (>20-fold) under H2O2 stress. [23] KatG, a catalase, and AphC, an NADH peroxidase, are activated, both of which directly reduce cellular peroxide. [24] [25] Several iron-related enzymes are also induced. Dps, a ferritin homolog, is most significantly upregulated (180-fold). Dps oxidizes cellular free Fe2+, and thus inhibits the Fenton chemistry and protects DNA from hydroxyl radical damage. [26] [27] [28] Similarly, in order to decrease free cellular Fe2+ concentration, a Ferric uptake regulator (Fur) protein is also induced, which serves as an inhibitor to iron import. Furthermore, MntH, a Mn importer, is induced, too. Mn2+ cannot be oxidized by H2O2 and is able to serve as an enzymatic cofactor. Thus, higher cellular Mn2+ concentration allows Mn2+ to substitute for Fe2+ and protect the iron-dependent enzymes from damage caused by the Fenton chemistry. [29] There are other regulon members that are induced significantly under H2O2 pressure. However, the functions of some are still unknown, while the others’ roles seem to be less relevant after mutation assay. Notably, there is no direct DNA or lipid repair gene regulated by OxyR.
OxyRs in Vibrio vulnificus
Vibrio vulnificus is a pathogenic Gram-negative bacterium. Although it is less infectious than other members in the Vibrio genus, it could result in severe and frequently fatal infections in immunocompromised individuals. The infection of V. vulnificus is usually related to consumption of raw shellfish, which bioaccumulate the bacteria via filter feeding. V. vulnificus can lead to two kinds of diseases: septicemia and tissue necrosis, both of which develop very quickly and could be fatal. [30] Since the immune system defends against this kind of pathogenic bacteria by generating ROS, thorough characterization of the mechanism of V. vulnificus under oxidative stress is crucial to develop a better clinical treatment for its infection. [31]
In previous research, two distinct peroxiredoxins (Prx), Prx1 and Prx2, have been identified in gram-negative facultative aerobe V. vulnificus [32] Prx1 is previously known as the homolog of NADH peroxidase AhpC in V. vulnificus. Prx1 is regulated by transcription factor OxyR in V. vulnificus and has a similar function as that found in E. coli. [33] [34] However, the discovery and characterization of Prx2 led researchers to hypothesize the presence of another OxyR-type transcription factor in V. vulnificus. Prx2 shares low amino acid sequence identity with Prx1 and other bacterial AhpC. Prx2 is able to sense H2O2 at a level that is much lower than what Prx1 is able to do. Since V. vulnificus is a facultative aerobe, two Prxs thus allow the bacteria to optimize the defensive process against different amounts of H2O2. However, it is not clear how V. vulnificus regulates the gene expression of the second Prx.
Kim et al. successfully identified and characterized OxyR2, a new OxyR-type transcription factor in V. vulnificus. Sequence analysis shows that OxyR2 shares 34% amino acid sequence identity to the first OxyR (thereafter OxyR1) and contains the two Cysteine residues located corresponding to the redox-sensitive ones in OxyR1. Similarly, the two Cysteine residues in OxyR2 are subject to H2O2 oxidation to form a reversible disulfide bond. The Prx2 expression is only induced by the oxidized form of OxyR2 in vivo. The minimum concentration of H2O2 required to oxidize OxyR2 is much lower than that of OxyR1, which explains the high sensitivity of OxyR2 and thus that of Prx2. Compared to OxyR1, the enhanced H2O2 sensitivity could be because of the fact that the microenvironment that the redox-active Cysteine residues locate are different. In OxyR1, Cys-199 is adjacent to a negatively charged Asp residue, while in OxyR2, the corresponding residue Cys-206 is adjacent to a positively charged Lys residue, which could enhance the nucleophilicity of Cys-206 by decreasing the thiol pKa, resulting in a higher reactivity to H2O2.
By possessing two OxyRs with different affinities to H2O2, V. vulnificus are able to fine-tune the detoxification of different amounts of H2O2 they may encounter in the natural environment. Such optimization could be an evolutionary advantage relevant to its pathogenesis. Furthermore, BLAST search reveals that there are many other bacteria containing homologs of both OxyR1 and OxyR2, suggesting that the coexistence of both OxyRs is widely distributed in bacteria.
From a medical perspective, the mutation on the key Cysteine residues in OxyR2 could result in a lower concentration of cellular lactate dehydrogenase in the infected host cells, which indicates a decreased cytotoxicity caused by the infection. In addition, during infection, the oxyR2 mutants showed a decreased growth rate compared to the wild type. This could be explained by the fact that the lack of OxyR2 caused decreased Prx2 in the bacteria, so they were not able to fight against the ROS generated by the immune system of the host cells. These results imply that OxyR2 might be important for the virulence of V. vulnificus. [35]
Defensive strategy for Gram-positive bacteria: PerR
H2O2 Sensory Mechanism of PerR
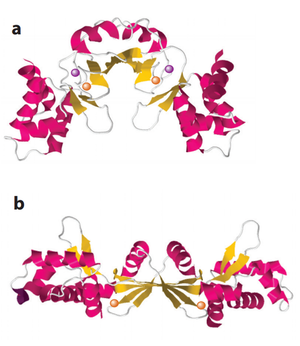
PerR is the common H2O2-inducible transcription factor in many Gram-positive bacteria such as Bacillus subtilis and Staphylococcus aureus. It belongs to the Fur protein family and uses a metal oxidation reaction to sense H2O2. PerR is a dimer and each subunit has a structural zinc-binding site that irreversibly binds to Zn2+. Each subunit also contains at least one regulatory metal binding site. The metal in the regulatory site varies between organisms, usually Fe2+ and/or Mn2+. [36] [37] When cellular H2O2 concentration increases, the regulatory metal will be oxidized. In contrast to the cysteine thiol-dependent sensory mechanism in OxyR, the oxidized metal causes rapid oxidation of histidine residues nearby and leads to a conformational change of the protein (Figure 4). [38] This conformational change causes a decreased binding affinity of PerR to DNA. Since PerR usually functions as a repressor, its detachment from DNA relieves the inhibition on the gene expression of the regulon members.
PerR in Bacillus subtilis
PerR in B. subtilis (PerRBS) is the most studied PerR in bacteria. In B. subtilis, the metal in the regulatory site is Fe2+. PerRBS may also bind Mn2+ as an alternative, which, however, as mentioned before, is not sensitive to H2O2. [39] PerRBS shows the same catalytic rate with H2O2 as that of OxyR in E. coli. [40] Compared to the reversible cysteine oxidation in OxyR, the oxidation of histidine residues in PerRBS seems to be irreversible, since there is no oxo-histidine reduction mechanism discovered yet. The PerRBS regulon overlaps a lot with that of OxyR in Gram-negative bacteria, such as KatA, a KatG homolog, MrgA, a Dps homolog, and a Fur protein. [41] However, contrary to OxyR, PerRBS regulon does not include the enzymes related to maintain thiol and to reduce disulfide bonds.[42]
PerR in Staphylococcus aureus
Staphylococcus aureus is a Gram-positive pathogenic bacterium. As an opportunistic pathogen, it is often found on the normal skin microflora and in the nasal passage and respiratory tract. However, on susceptible individuals, it is the most common cause of mild skin infections as well as often-fatal infections including sepsis and toxic shock syndrome. [43] Current therapeutics of S. aureus infection are increasingly complicated because of the emergence and prevalence of methicillin-resistant S. aureus (MRSA) across the world. [44] It has been shown that in the infected organisms, their neutrophils use phagosomes containing ROS to defend against S. aureus. [45] Therefore, in order to develop enhanced treatments to S. aureus infection, it is important to characterize the mechanism(s) that S. aureus uses to defend against oxidative stress.
PerR has been previously identified as the major transcription factor on S. aureus to respond to oxidative burst. [46] Ji et al. characterized the PerR in S. aureus (PerRSA) and showed that it is an H2O2-inducible translation factor using the same sensory mechanism as B. subtilis. Together with PerR found in other bacteria, Ji et al. suggest that PerR and PerR-type transcription factors may be a conserved mechanism for Gram-positive bacteria against H2O2 pressure. However, under the same H2O2 pressure, the expression levels of PerR regulon were higher in S. aureus expressing PerRSA than those expressing PerRBS, which suggests that PerRSA was more sensitive to H2O2 oxidation and thus more resistant to H2O2.
Furthermore, Ji et al. used Caenorhabditis elegans as the model to study the relationship between the H2O2 sensitivity of PerR and the pathogenesis of S. aureus. They observed that the S. aureus expressing PerRBS killed C. elegans faster than the PerRSA. This suggested that the lower susceptibility of PerR to H2O2 oxidation actually increases the virulence of S. aureus. [47]
Defending against superoxide (O2-)
Why is O2- poisonous?
Since O2- is a negatively charged molecule, it cannot cross the cell membrane. [48] Therefore, the cellular O2- is formed endogenously. O2- may come from either adventitious autoxidation of flavoproteins or redox-cyclic organic chemicals. The redox-cyclic organic compounds can cross the cell membrane, steal electrons from redox enzymes in the target cell and transfer to molecular oxygen, resulting in a toxic level of O2- in the cell. As a result, some bacteria and plants excrete redox-cyclic organic chemicals, such as quinones and phenazines, as extracellular electron carriers and as their weapons against microflora. For example, in the lungs of cystic fibrosis (CF) patients, Hunter et al. have found that the dominant bacteria Pseudomonas aeruginosa produces phenazines in order to both damage the host epithelial cells and hinder the growth of other bacteria. Higher phenazine concentration would both decrease microfloral complexity and accelerate the CF disease progression. [49]
Toxic level of O2- in the cell could interfere with the function of [4Fe-4S] cluster in many important enzymes by oxidation, such as aconitase and fumarases in the TCA cycle and 6-phosphogluconate dehydrogenase in the pentose phosphate pathways. Such enzyme deficiency would lead to certain amino acid auxotrophies including branched-chain, aromatic and sulfurous amino acids. Bacteria with such a deficiency cannot grow unless supplemented with those amino acids. [50] In order to survive under the oxidative burst, bacteria such as E. coli will increase their cellular superoxide dismutase (SOD), which is mediated by transcription factors SoxR and SoxS in a two-stage manner. [51] SoxR is widely distributed in Actinobacteria, Enterobacteriales, Pseudomonadaceae and α-, β-, γ-, and δ-proteobacteria. [52] SoxR contains a [2Fe-2S] metal center in its active site that is sensitive to O2-. Once activated, SoxR would trigger the transcription of soxS gene, increasing the cellular SoxS concentration. SoxS then would initiate the transcription of O2- defensive genes. The gene products of SoxRS are very diverse and overlap with those regulated by OxyR. The regulon members include genes encoding for scavenging enzymes (SOD and AhpCF) and superoxide-resistant dehydratases (Fumarase C and Aconitase A). The latter ones are used to substitute for the endogenous redox-sensitive enzymes to restore the important cellular function including the TCA cycle and the pentose phosphate pathway. [53]
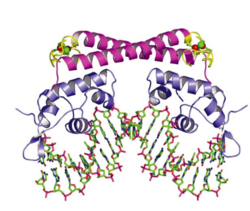
O2- Sensory Mechanism of SoxR
In E. coli, each subunit in the dimeric SoxR protein contains one redox-sensitive [2Fe-2S]+ metal cluster and one DNA-binding motif. When the bacteria are exposed to redox-cycling chemicals, the [2Fe-2S]+ metal clusters are oxidized in a reversible reaction. This oxidation causes a conformational change of SoxR and therefore twists the DNA binding on SoxR, sliding the promoter of soxS to a place where RNA polymerase can efficiently bind and start transcription (Figure 5). [54] Once the oxidants are cleared away, the oxidized SoxR is reduced back to its original state catalyzed by Rsx/Rse enzymes probably using NADPH as an electron donor, [55] and the SoxS is quickly degraded via proteolysis and thus stops the transcription of the whole SoxRS regulon. [56]
SoxR in Pseudomonas aeruginosa
The opportunistic human pathogen P. aeruginosa can cause various and often-fatal diseases on susceptible individuals. As mentioned above, it is the dominant bacterium found in patients with cystic fibrosis lung, damaging the pulmonary epithelial cells and outcompeting other microbes in the environment by producing phenazine, a redox-cycling compound. [57] As a Gram-negative bacterium, both SoxR and OxyR homologs have been previously found in P. aeruginosa. OxyR homolog in P. aeruginosa functions similarly to the one found in E. coli, which is mainly against H2O2 pressure. [58] Since its virulence is closely related to the oxidative phenazine generated endogenously, it is important to find out the mechanism of how the bacterium protects itself, and the relationship between phenazine production and SoxR transcriptional regulation.
Surprisingly, no SoxS was identified in P. aeruginosa as the secondary transcription factor downstream of SoxR. Indeed, SoxSs are strictly expressed in the family Enterobacteriaceae. [59] Therefore, the primary function of SoxR in P. aeruginosa may not be against O2- like the one in E. coli. Palma et al. have characterized the transcriptome profile under O2- stress and elucidated the members in the SoxR regulon and their functions in P. aeruginosa. Six genes have been identified in the regulon, including a gene encoding putative monooxygenase, a gene for probable efflux pump and a 4 gene mexGHI-ompD operon for multidrug efflux pump. Mutation assays suggest that SoxR in P. aeruginosa is not the major transcription factor against oxidative stress (both H2O2 and O2-). It is not related to antibiotic resistance, either. However, SoxR is very important in the virulence of P. aeruginosa pulmonary infection in a mouse model. P. aeruginosa strains with deletion of soxR became non-virulent to the mice, while the wild-type killed the mice after 3 days. Although the specific mechanism that SoxR may use for virulence is still unclear, Palma et al. suggested that the MexGHI-ompD system could be relevant. Since MexGHI-ompD is important in quorum-sensing signal homeostasis, O2--induced expression of it may result in a global physiological alternative and thus a change in virulence. [60]
Conclusion
OxyR, PerR and SoxR are three extensively studied bacterial transcription factors that are sensitive to two of the most common ROS: peroxide and superoxide. There is some overlapping in members of the regulon of these three important transcription factors. However, variations in different bacteria species are also identified, such as the sensory mechanisms. Additionally, there are other bacterial transcription factors identified that bacteria use to sense and defend against other ROS, including organic hydroperoxides, hypochlorous acid, and disulfide stress. Thus, the word "oxidative stress" is very broad, as different ROS have distinct effects on the transcriptional regulatory system on bacteria. Due to its crucial role in bacterial virulence and antibiotic resistance, a thorough understanding of the complex system that the bacteria, especially the pathogens, defend themselves against ROS is required for better strategies for us to fight against them.
References
- ↑ Halliwell (2006): Reactive Species and Antioxidants. Redox Biology Is a Fundamental Theme of Aerobic Life. Plant Physiology 141(2):312–322.
- ↑ Seaver and Imlay (2004): Are Respiratory Enzymes the Primary Sources of Intracellular Hydrogen Peroxide? The Journal of Biological Chemistry, 279:48742-48750.
- ↑ Apostol et al. (1989): Rapid Stimulation of an Oxidative Burst during Elicitation of Cultured Plant Cells: Role in Defense and Signal Transduction—Commentary. Plant Physiology, 90:109–116.
- ↑ Yang et al.(2013): Reactive oxygen species in the immune system. International Reviews of Immunology 32(3):249-70.
- ↑ Imlay (2015): Transcription factors that defend bacteria against reactive oxygen species 2015. Annual Review of Microbiology 69:93–108.
- ↑ Choi et al. (2013): Structural Basis of the Redox Switch in the OxyR Transcription Factor. Cell 105(1):103-113.
- ↑ Zheng et al. (1998): DNA Microarray-Mediated Transcriptional Profiling of the Escherichia coli Response to Hydrogen Peroxide. Journal of Bacteriology 183(15): 4562-4570.
- ↑ Kim et al. (2014): Distinct characteristics of OxyR2, a new OxyR-type regulator, ensuring expression of Peroxiredoxin 2 detoxifying low levels of hydrogen peroxide in Vibrio vulnificus. Molecular Microbiology 93(5):992–1009.
- ↑ Ji et al. (2015): Staphylococcus aureus PerR Is a Hypersensitive Hydrogen Peroxide Sensor using Iron-mediated Histidine Oxidation. The Journal of Biological Chemistry 290:20374-20386.
- ↑ Lee & Helmann (2006): The PerR transcription factor senses H2O2 by metal-catalysed histidine oxidation. Nature 440:363-367.
- ↑ Greenberg et al. (1990): Positive control of a global antioxidant defense regulon activated by superoxide-generating agents in Escherichia coli. PNAS 87: 6181-6185.
- ↑ Tsaneva and Weiss (1990): soxR, a locus governing a superoxide response regulon in Escherichia coli K-12. Journal of Bacteriology 172(8):4197-205.
- ↑ Blanchard et al. (2007): Rapid Changes in Gene Expression Dynamics in Response to Superoxide Reveal SoxRS-Dependent and Independent Transcriptional Networks. PLoS ONE 2(11): e1186.
- ↑ Mehdy (1994): Active Oxygen Species in Plant Defense against Pathogens. Plant Physiology 105(2):467-472.
- ↑ Bedard and Krause (2007): The NOX Family of ROS-Generating NADPH Oxidases: Physiology and Pathophysiology. Physiological Reviews 87(1):245-313.
- ↑ Seki et al. (2004): Hydrogen Peroxide Production in Streptococcus pyogenes: Involvement of Lactate Oxidase and Coupling with Aerobic Utilization of Lactate. Journal of Bacteriology 186(7):2046–2051.
- ↑ Machlin and Bendich (1987): Free radical tissue damage: protective role of antioxidant nutrients. The FASEB Journal 1(6):441-445.
- ↑ Hogg et al. (2005): Bumps in the road: how replicative DNA polymerases see DNA damage. Current opinion in structural biology 15(1): 86-93.
- ↑ Imlay et al. (1988): Toxic DNA damage by hydrogen peroxide through the Fenton reaction in vivo and in-vitro. Science 240(4852): 640-642.
- ↑ Choi et al. (2013): Structural Basis of the Redox Switch in the OxyR Transcription Factor. Cell 105(1):103-113.
- ↑ Jo et al. (2014): Structural details of the OxyR peroxide-sensing mechanism. PNAS 112(20):6443–6448.
- ↑ Choi et al. (2013): Structural Basis of the Redox Switch in the OxyR Transcription Factor. Cell 105(1):103-113.
- ↑ Zheng et al. (1998): DNA Microarray-Mediated Transcriptional Profiling of the Escherichia coli Response to Hydrogen Peroxide. Journal of Bacteriology 183(15): 4562-4570.
- ↑ Heym et al. (1995): Missense mutations in the catalase‐peroxidase gene, katG, are associated with isoniazid resistance in Mycobacterium tuberculosis. Molecular microbiology 15(2):235-245.
- ↑ Ellis & Poole (1997): Roles for the two cysteine residues of AhpC in catalysis of peroxide reduction by alkyl hydroperoxide reductase from Salmonella typhimurium. Biochemistry 36(43):13349-13356.
- ↑ Grant et al.(1998): The crystal structure of Dps, a ferritin homolog that binds and protects DNA. Nature Structural & Molecular Biology 5(4): 294-303.
- ↑ Ilari et al. (2005): The unusual intersubunit ferroxidase center of Listeria innocua Dps is required for hydrogen peroxide detoxification but not for iron uptake. A study with site-specific mutants. Biochemistry 44(15): 5579-5587.
- ↑ Chiancone & Ceci (2010): The multifaceted capacity of Dps proteins to combat bacterial stress conditions: detoxification of iron and hydrogen peroxide and DNA binding. Biochimica et Biophysica Acta (BBA)-General Subjects, 1800(8): 798-805.
- ↑ Kehres et al. (2000): The NRAMP proteins of Salmonella typhimurium and Escherichia coli are selective manganese transporters involved in the response to reactive oxygen. Molecular microbiology 36(5): 1085-1100.
- ↑ Strom & Paranjpye (2000): Epidemiology and pathogenesis of Vibrio vulnificus. Microbes and infection 2(2): 177-188.
- ↑ Chung et al. (2010): RtxA1-induced expression of the small GTPase Rac2 plays a key role in the pathogenicity of Vibrio vulnificus. Journal of Infectious Diseases, 201(1): 97-105.
- ↑ Bang et al. (2012): Distinct characteristics of two 2-Cys peroxiredoxins of Vibrio vulnificus suggesting differential roles in detoxifying oxidative stress. Journal of Biological Chemistry 287(51): 42516-42524.
- ↑ Baek et al. (2009): Identification of the Vibrio vulnificus ahpCl gene and its influence on survival under oxidative stress and virulence. The Journal of Microbiology 47(5):624-632.
- ↑ Kong et al. (2004): Role of catalase and oxyR in the viable but nonculturable state of Vibrio vulnificus. FEMS microbiology ecology 50(3):133-142.
- ↑ Kim et al. (2014): Distinct characteristics of OxyR2, a new OxyR-type regulator, ensuring the expression of Peroxiredoxin 2 detoxifying low levels of hydrogen peroxide in Vibrio vulnificus. Molecular Microbiology 93(5):992–1009.
- ↑ Traoré et al. (2006): Crystal structure of the apo‐PerR‐Zn protein from Bacillus subtilis. Molecular microbiology 61(5): 1211-1219.
- ↑ Jacquamet et al. (2009): Structural characterization of the active form of PerR: insights into the metal‐induced activation of PerR and Fur proteins for DNA binding. Molecular microbiology 73(1): 20-31.
- ↑ Lee & Helmann (2006): Biochemical characterization of the structural Zn2+ site in the Bacillus subtilis peroxide sensor PerR. Journal of Biological Chemistry 281(33): 23567-23578.
- ↑ Chen et al.(1995): Coordinate regulation of Bacillus subtilis peroxide stress genes by hydrogen peroxide and metal ions. Proceedings of the National Academy of Sciences 92(18): 8190-8194.
- ↑ Lee & Helmann (2006): Biochemical characterization of the structural Zn2+ site in the Bacillus subtilis peroxide sensor PerR. Journal of Biological Chemistry 281(33): 23567-23578.
- ↑ Bsat et al. (1998): Bacillus subtilis contains multiple Fur homologues: identification of the iron uptake (Fur) and peroxide regulon (PerR) repressors. Molecular microbiology 29(1): 189-198.
- ↑ Imlay (2015): Transcription factors that defend bacteria against reactive oxygen species 2015. Annual Review of Microbiology 69:93–108.
- ↑ Lowy (1998): Staphylococcus aureus infections. New England journal of medicine 339(8): 520-532.
- ↑ Klevens et al.(2007): Invasive methicillin-resistant Staphylococcus aureus infections in the United States. Jama 298(15): 1763-1771.
- ↑ Spaan et al. (2013): Neutrophils Versus Staphylococcus aureus: A Biological Tug of War. Annual review of microbiology 67: 629-650.
- ↑ Horsburgh et al. (2001): PerR controls oxidative stress resistance and iron storage proteins and is required for virulence in Staphylococcus aureus. Infection and immunity 69(6): 3744-3754.
- ↑ Ji et al. (2015): Staphylococcus aureus PerR Is a Hypersensitive Hydrogen Peroxide Sensor using Iron-mediated Histidine Oxidation. The Journal of Biological Chemistry 290:20374-20386.
- ↑ Korshunov & Imlay (2002): A potential role for periplasmic superoxide dismutase in blocking the penetration of external superoxide into the cytosol of Gram‐negative bacteria. Molecular microbiology 43(1): 95-106.
- ↑ Hunter et al. (2012): Phenazine content in the cystic fibrosis respiratory tract negatively correlates with lung function and microbial complexity. American journal of respiratory cell and molecular biology 47(6): 738-745.
- ↑ Imlay (2013): The molecular mechanisms and physiological consequences of oxidative stress: lessons from a model bacterium. Nature Reviews Microbiology 11(7): 443-454.
- ↑ Nunoshiba et al. (1992): Two-stage control of an oxidative stress regulon: the Escherichia coli SoxR protein triggers redox-inducible expression of the soxS regulatory gene. Journal of Bacteriology 174(19): 6054-6060.
- ↑ Dietrich et al. (2008): Redox-active antibiotics control gene expression and community behavior in divergent bacteria. Science 321(5893): 1203-1206.
- ↑ Pomposiello & Demple (2001): Redox-operated genetic switches: the SoxR and OxyR transcription factors. Trends in biotechnology 19(3): 109-114.
- ↑ Watanabe et al. (2008): Crystal structure of the 2Fe-2S oxidative-stress sensor SoxR bound to DNA. Proceedings of the National Academy of Sciences, 105(11): 4121-4126.
- ↑ Koo et al. (2003): A reducing system of the superoxide sensor SoxR in Escherichia coli. The EMBO journal 22(11): 2614-2622.
- ↑ Griffith et al. (2004): Proteolytic degradation of Escherichia coli transcription activators SoxS and MarA as the mechanism for reversing the induction of the superoxide (SoxRS) and multiple antibiotic resistance (Mar) regulons. Molecular microbiology, 51(6), 1801-1816.
- ↑ Hunter et al. (2012): Phenazine content in the cystic fibrosis respiratory tract negatively correlates with lung function and microbial complexity. American journal of respiratory cell and molecular biology 47(6): 738-745.
- ↑ Ochsner et al. (2000): Role of the Pseudomonas aeruginosa oxyR-recG operon in oxidative stress defense and DNA repair: OxyR-dependent regulation of katB-ankB, ahpB, andahpC-ahpF. Journal of bacteriology 182(16): 4533-4544.
- ↑ Dietrich et al. (2008): Redox-active antibiotics control gene expression and community behavior in divergent bacteria. Science 321(5893): 1203-1206.
- ↑ Palma et al. (2005) Pseudomonas aeruginosa SoxR does not conform to the archetypal paradigm for SoxR-dependent regulation of the bacterial oxidative stress adaptive response. Infection and immunity 73(5): 2958-2966.
Authored for BIOL 238 Microbiology, taught by Joan Slonczewski, 2016, Kenyon College.
