Bioremediation
Introduction
Bioremediation refers to the use of microorganisms to degrade contaminants that pose environmental, and especially human risks. It has become an accepted remedy for cleaning polluted soil and water due to its safety and convenience. Bioremediation allows scientists to concentrate clean-up efforts at the site of contamination. [1] Bioremediation processes typically involve many different microbes acting in parallel or sequence to complete the degradation process. The ability of microbes to degrade a vast array of pollutants makes bioremediation a widely applicable technology that can applied in different soil conditions [3].
A widely used approach to bioremediation involves stimulating a group of organisms in order to shift the microbial ecology toward the desired process. This is termed "Biostimulation." Biostimulation can be achieved through changes in pH, moisture, and aeration. One of the most common approaches to bioremediation involves in-situ addition of nutrients and oxygen. The other widely used approach is termed "Bioaugmentation" where organisms selected for high degradation abilities are used to inoculate the contaminated site [3]. These two approaches are not mutually exclusive- they can be used simultaneously. Bioreactors can also be employed for remediation. In such cases, soil and groundwater from the contaminated site are transported to the reactor, where conditions favorable for biological reactions are enhanced [5].
Example Microorganisms
Pseudomonas putida
Pseudomonas putida is a gram-negative soil bacterium that is involved in the bioremediation of toulene, a component of paint thinner. It is also capable of degrading naphthalene, a product of petroleum refining, in contaminated soils. [2]
Nitrosomonas europaea, Nitrobacter hamburgensis, and Paracoccus denitrificans
Industrial bioremediation is used to clean wastewater. Most treatment systems rely on microbial activity to remove unwanted mineral nitrogen compounds (i.e. ammonia, nitrite, nitrate). The removal of nitrogen is a two stage stage process than involves nitrification and denitrification (see Nitrogen cycle including GHG). During nitrification, ammonium is oxidized to nitrite by organisms likeNitrosomonas europaea.The, nitrite is further oxidized by microbes like Nitrobacter hamburgensis.
In anaerobic conditions, nitrate produced during ammonium oxidation is used as a terminal electron acceptor by microbes likeParacoccus denitrificans[2]. The result is dinitrogen gas. Through this process, ammonium and nitrate, two pollutants responsible for eutrophication in natural waters, are remediated.
Phanerochaete chrysosporium
The lignin-degrading white rot fungus, Phanerochaete chrysosporium, exhibits strong potential for bioremediation of: pesticides, polyaromatic hydrocarbons, PCBs, dioxins, dyes, TNT and other nitro explosives, cyanides, azide, carbon tetrachloride, and pentachlorophenol. White rot fungi degrade lignin with nonselective extracellular peroxidases, which can also facilitate the degradation of other compounds containing similar structure to lignin within the proximity of the enzymes released [6].
Deinococcus radiodurans
Deinococcus radiodurans is a radiation-resistant extremophile bacterium that is genetically engineered for the bioremediation of solvents and heavy metals. An engineered stain of Deinococcus radiodurans has been shown to degrade ionic mercury and toluene in radioactive mixed waste environments [7].
Methylibium petroleiphilum
Methylibium petroleiphilum(formally known as PM1) is a bacterium is capable of methyl tert-butyl ether (MTBE) bioremediation. PM1 degrades MTBE by using the contaminant as the sole carbon and energy source [8].
Example Pollutants
Pollutants found in soils present a variety of different human health risks including direct toxicity, as well as bioaccumulation in plant and animal tissue eventually consumed by humans. Pollutants that are being studied for bioremediation potential are listed below. The remediation of some of these pollutants will be discussed in greater depth in the following sections.
Petroleum byproducts
BTEX - benzene, toluene, ethylbenzene, and xylene - are byproducts of petroleum products. The biodegradability of these compounds is relatively well known and remediation can be achieved by creating favorable conditions for BTEX degrader's growth. PAH - Polycyclic aromatic compounds remain on the soil surface and are harder to degrade than BTEX [3].
Methyl tert-butyl ether
MTBE is a gasoline additive introduced to replace lead. MTBE raises the oxygen content of fuel, allowing for more complete combustion and less emissions. MTBE, however, is highly soluble, does not adsorb well in soil and can therefore move quickly through soil and into groundwater [4].
Polychlorinated bhiphenols
PCBs are used in industrial applications, are very recalcitrant, and many are known carcinogens.
Chlorinated solvents
Chlorinated solvents are used extensively as cleaning agents. Plumes have been found to contaminate groundwater below dry cleaners in many places, including Davis, Ca. Many chlorinated solvents are carcinogenic. TCE can be degraded to vinyl chloride under anaerobic conditions. Vinyl chloride, in tern, needs different conditions to transform, and this should be seriously considered due to its high toxicity [3].
Polynuclear aromatic compounds
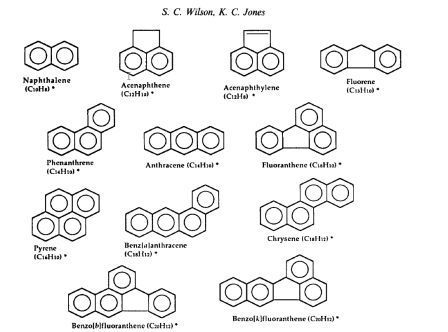
PAHs are found in high concentrations at industrial sites especially sites that use or process petroleum products. The are considered carcinogens and mutanogens, and are very recalcitrant, pervading for many years in the natural environment.
Other contaminants include residuals from flares (perchlorate) and explosives (TNT, RDX); metals (chromium, lead); plutonium and uranium; polynuclear aromatic compounds; potassium and nitrogen. Much of the high levels of these contaminants found in nature is a result of human activity [3]
Bioremediation Applications
Exxon Valdez Oil Spill in Prince William Sound
Bioremediation was employed to treat the 1989 Exxon Valdez oil spill in Prince William Sound, Alaska. Hydrocarbon degrading microbes exist in marine systems because natural sources of hydrocarbon exists as a result of geological seeps and other sources. During the Exxon cleanup effort, the activity of these organisms was enhanced through the addition of nitrogen and phosphorus to oil laden beaches [9]. This is an example of bio-stimulation.
Metabolic Pathways
Microorganisms use a wide range of metabolic pathways to harvest energy from their environment. In some cases, pollutants serve as the carbon and energy source for microbial growth, while in other cases, pollutants serve as the terminal electron acceptor (ex. perchlorate degradation). This manifests itself in the diverse ability of microbes to transform and degrade toxic molecules. The degradation pathways for a few of the pollutants listed above are explored.
Polychlorinated Biphenyls PCBs
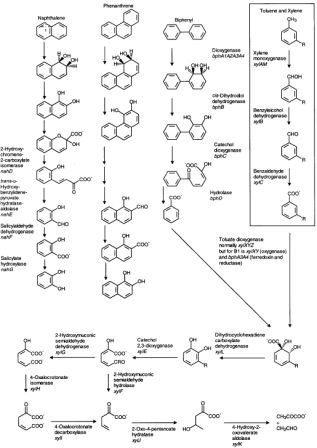
Metabolism of polychlorinated biphenyls is generally through to proceed through the addition of two oxygens to the aromatic ring, followed by ring cleavage as seen the the metabolic pathways diagram. Energy is obtained through the oxidation of these large hydrocarbons [12].
Polynuclear aromatic compounds (PAHs)
Examples of PAHs are seen below:
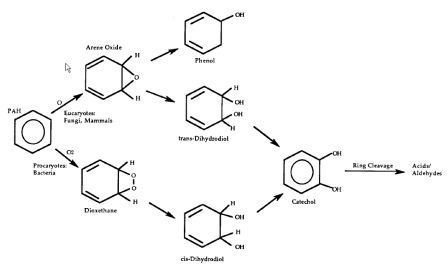
PHAs in contaminated soils can be treated with bioremediation. The oxidation of PAH involves oxygenases (monooxygenases and dioxygenases). Fungi complete the process by adding an oxygen to the substrate PAH to form arene oxides and then enzymatically adding water to form trans-dihydrodiols and phenols. Bacteria mainly use dioxygenases, adding two oxygens to the substrate and the further oxidizing it to dihydrodiols and dihydroxy products. Ring oxidation is the rate limiting step in the reaction, and subsequent reactions occur fairly quickly, yielding the typical metabolic intermediate Catechol found in Lignin degradation as well as Gentisic and Protocatechuic Acids (see diagram below) [5].
Intermediate metabolites degrade further through ortho and meta ring cleavage to produce succinic, fumaric, pyruvic, and acetic acids and acetyl-CoA, which are shunted into major metabolic and anabolic pathways [11]. The byproducts of these reactions are carbon dioxide and water. The breakdown of PAHs can occur when microorganisms use PAH as their sole energy and carbon source and also through the co-metabolisms process. Co-metabolism refers to when an enzyme directed at another compound also degrades PHA. This has been shown to be an important phenomenon in breaking down larger aromatic chains, by does not directly lead to complete oxidation to carbon dioxide [5].
Monitoring
To monitor the bioremedation potential of a soil one can probe for the existence of specific degradation pathways in the soil community or monitor for specific enzymes involved in the process. There are two common ways to test for functional genes involved in the degradation of a compound. First, specific DNA hybridization probes can be used to indicate potential for the organisms to degrade the desired compound. Second, specific RNA hybridization probes are used to indicate the expression of the functional genes in the environment[3].
The actual change in pollutant concentration or degradation byproducts can also be monitored to determine the amount of pollutant removal. To determine if the degradation of a desired compound is the result of abiotic or biotic activity, controlled laboratory experiments are used. The concentration of a pollutant in a non-sterile microcosm containing soil from the environment of interest is compared to a sterile control. The sterile control shows the non-biological contribution to the disappearance of the pollutant due to, for example, adsorption to clay particles or precipitation. The non-sterile microcosm simulates the microbial contribution to the degradation of the pollutant in the natural environment, but also includes other abiotic mechanisms. The microbial contribution to pollutant disappearance is the difference between removal in the biologically active bottle and removal in the sterile control. This helps to quantify whether the disappearance of the pollutant is the result of biological or non-biological mechanisms. [3]
Current Research
Pseduomonas putida
Pseudomonas putida has been found to be useful in the detection of certain chemicals, such as land mines. On the grand scale, a linkage between the bacteria's ability to degrade TNT and the explosive compound found in land mines has inspired research to utilize P. putida as a way of detecting land mines from soil content. TSCA Experimental Release Application Approved for Pseudomonas putida Strains
Nitrosomonas europaea
One possible treatment for the purification of water has been the use of Trihalomethanes or THM's. Recent studies have linked these four chemicals, tricholormethane or chloroform, bromomethane, dibromomethane and dichlorobromomethane have been linked to colon cancer. [12] Because of its nitrogen oxidizing properties, Nitrosomonas Europea has been studied under ammonia rich conditions and THM rich conditions, recognized as limiting reactants in the conversion of ammonia. [13]
Methylibium petroleiphilum
A motile, gram-negative facultative anaerobic bacterium, [Methylibium petroleiphilum] has been isolated because its ability to completely mineralize methyl tert-butyl ether (MTBE), a gasoline additive. Methylibium petroleiphilum is capable of consuming a diverse range of gasoline derivatives as its sole carbon source, including: methanol, ethanol, toluene, benzene, ethylbenzene, and dihydroxybenzenes. Optimal growth of M. petroleiphilum occurs at the soil subsurface with pH of 6.5 and 30°C. The upper temperature limit of this bacterium is 37°C. [14]
References
1. States Environmental Protection Agency, "A Citizen's Guide to Bioremediation" 2001.
2. Nitrification and Denitrification Wastewater Treatment. No. 5536407. 16 July 1996.
3. Sylvia, D. M., Fuhrmann, J.F., Hartel, P.G., and D.A Zuberer (2005). "Principles and Applications of Soil Microbiology." New Jersey, Pearson Education Inc.
4. States Environmental Protection Agency, "MTBE," 2007
5. Wilson, S. C., and Kevin C. Jones (1993). "Bioremediation of Soil Contaminated with Polynuclear Aromatic Hydrocarbons (PHAs): A review." Environmental Pollution. 81: 229-49.
10. Pritchard, PH. 1991. "Bioremediation as a technology: experiences with the Exxon Valdez oil spill." Journal of Hazardous Materials 28:115-130.
11. Scow, Kate. "Lectures in Soil Microbiology." UC Davis, Winter 2008.
12. Oram, Brian. "Disinfection By-Products Trihalomethanes." Wilkes University, 2003
Edited by student of Kate Scow