Candidatus Chloracidobacterium thermophilum
Classification
Domain (Bacteria); Super Phylum (Fibrobacteres/Acidobacteria group); Phylum (Acidobacteria); Class (Acidobacteria); Order (Acidobacteriales); Family (Acidobacteriaceae); Genus (Candidatus Chloracidobacterium)
Species
Candidatus Chloracidobacterium thermophilum
Description and Significance
A novel photosynthesizing bacterium named as Candidatus Chloracidobacterium thermophilum (Cab. thermophilum) was recently (2007) discovered in Yellowstone’s hot springs by a collaborative research between The Pennsylvania State University and Montana State University. The research was funded by National Science Foundation, Department of Energy, and the NASA Exobiology Program.[1]
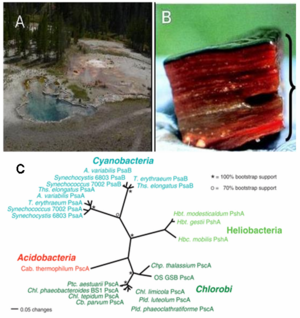
The researchers have found the genetic evidence of this novel bacterium in three hot springs (Mushroom Spring, Octopus Spring, Green Finger Pool) in Yellowstone National Park (Figure 1A). It resides in the phototrophic microbial mats at moderately high temperature (50-66 degree celsius) and in alkaline environment (pH 8.5) (Figure 1B). Based on phylogenetic analysis (16S rRNA), it is the member of the Acidobacteria phylum, which did not previously contain any phototrophs. In addition, the analysis showed that microorganisms closely related to Cab. thermophilum also exist in hot springs in Tibet and Thailand.[1] The importance of this discovery is reflected by 'This is only the third time in the last 100 years that a new group of phototrophs has been discovered'.[2]
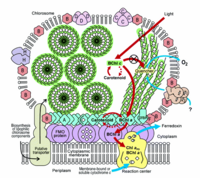
Until this discovery, chlorophyll-based phototrophy was only known to occur in only five groups or phyla of microorganisms (Cyanobacteria, Chloroflexi, Chlorobi, Proteobacteria and Firmicutes).[3] These phototrophic microorganisms contain specific types of photosynthetic pigments (chlorophylls and bacteriochlorophylls) to absorb specific wavelengths of light and differ in the type of molecular functions for the conversion of light into chemical energy (Figure 2). Different groups of phototrophic microorganisms contain different types of chlorophylls and bacteriochlorophylls.[4] For example Bacteriochlorophyll a-Purple bacteria, Cab. thermophilum :Bacteriochlorophyll b-Purple bacteria :Bacteriochlorophyll c-Green sulfur bacteria,Chloroflexi,Cab.thermophilum :Bacteriochlorophyll d-Green sulfur bacteria :Bacteriochlorophyll e-Green sulfur bacteria :Bacteriochlorophyll g-Heliobacteria :Chlorophyll a- Cyanobacteria, green sulfur bacteria, Cab.thermophilum :Chlorophyll b- Cyanobacteria.
Cab.thermophilum has light harvesting antennae (chlorosomes), Fenna-Matthews-Olson (FMO) protein, and type 1 reaction centers: each chlorosome can contain up to 250,000 bacteriochlorophyll molecules. Cab.thermophilum synthesizes bacteriochlorophylls a and c (BChl) under oxic conditions (photoheterotropically) unlike the anoxic conditions in other phototrophs. Characterization of a model green sulfur bacterium Chlorobium tepidum showed that BChl c molecules forms large aggregates by self-organization due to the unique molecular structure of the bacteriochlorophylls. The formation of large molecular aggregates of BChl c is independent of any protein scaffold. These protein-independent and self-organization properties lead to the formation of very large amounts of BChl c (eg. in Green sulfur bacteria, 25-30% of the total cellular carbon is utilized in the formation of BChl c) needed for cell growth at very low light environment. Genome sequencing of a phototroph from the Chlorobi phylum, Chlorobium tepidum has shown the involvement of ten proteins required for formation of the chlorosome envelope and has led to the identification of the enzymes required for the formation of BChl a and c molecules and a photosynthetic apparatus model is proposed (Figure 3).[5,6] Recent genome sequencing studies on Cab. thermophilum indicate that the chlorosomes contain fewer proteins but the pathways for the synthesis of BChl a and BChl c are very similar except for the formation of the isocyclic ring of the chlorin/bacteriochlorin moiety. [Personal communication with Dr. Donald A. Bryant]
The biochemical analysis of Cab.thermophilum chlorosomes has shown the presence of ketocarotenoids, similiar to Cyanobacteria, which may have the role in photoprotection under the oxic conditions. Further ongoing characterization (biochemical and genome sequencing) at The Pennsylvania State University, Montana State University, and Los Alamos National Laboratory will elucidate the role and diversity of this novel bacterium.
Genome Structure
Based on the recently completed genome sequencing of Cab. thermophilum, it is 3.7 Mb and expected to be divided unequally between two circular molecules. The smaller is about 0.5 to 0.6 Mb--the exact size is still uncertain. [Personal communication with Dr. D. A. Bryant]
Cell Structure, Metabolism and Life Cycle
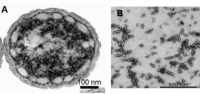
Cab. thermophilum is a Gram-negative, aerobic, rod-shaped bacterium. It is 2.5 microns long and about 0.7 microns in diameter (Figure 4). It is an anoxygenic photoheterotroph, which grows in the presence of light, organic carbon, and oxygen at moderate temperature (50-66 degree C) and alkaline (pH 8.5) environment. Cab. thermophilum synthesizes chlorosomes, Fenna-Matthews-Olson (FMO) protein, and type 1 reaction centers (PscA: type 1 reaction center protein). The Fenna-Matthews-Olson (FMO) protein-chlorophyll complex mediates the excitation energy transfer from the light-harvesting chlorosomes to the membrane embedded type 1 reaction center. [8]. The bacteriochlorophylls (BChl a and c) present in the type 1 reaction center absorb light and promotes the electron transfer within the BChls.[9]
Ecology and Pathogenesis
The researchers have found the genetic evidence of this novel bacterium in three hot springs (Mushroom Spring, Octopus Spring, Green Finger Pool) in Yellowstone National Park (Figure 1A). It resides in the phototrophic microbial mats at moderately high temperature (50-66 degree celsius) and in alkaline environment (pH 8.5) (Figure 1B).In addition, 16S rDNA analysis showed that microorganisms closely related to Cab. thermophilum also exist in hot springs in Tibet and Thailand.[1] Cab. thermophilum did not show any sign of pathogenicity to microbes or humanbeing so far.
References
[1] Bryant, D. A, Costas, A. M. G, Maresca, J. A, Chew, A. M. G, Klatt, C. G, Bateson, M. M, Tallon, L. J, Hostetler, J, Nelson, W. C, Heidelberg, J. F, and Ward, D. M. "Candidatus Chloracidobacterium thermophilum: An Aerobic Phototrophic Acidobacterium". " Science". 2007. Volume 317. p. 523 - 526[2]
[2] http://www.bmb.psu.edu/faculty/bryant/lab/Project/Acido/index.html
[3] Bryant, D. A and Frigaard, N.-U. "Prokaryotic photosynthesis and phototrophy illuminated". "Trends Microbiology". 2006. Volume 14. p. 488-496 [3]
[4] http://en.wikipedia.org/wiki/Bacteriochlorophyll
[5] Frigaard, N.-U. and Bryant, D. A. "Seeing green bacteria in a new light: genomics-enabled studies of the photosynthetic apparatus in green sulfur bacteria and filamentous anoxygenic phototrophic bacteria". "Archives of Microbiology". 2004. Volume 182. p. 265-276 [4]
[6] Frigaard, N.-U., Voigt, G. D., and Bryant, D. A. "Chlorobium tepidum mutant lacking bacteriochlorophyll c made by inactivation of the bchK gene, encoding bacteriochlorophyll c synthase". "Journal of Bacteriology". 2002. Volume 184. p. 3368-3376 [5]
[7] Li, H. Frigaard N.-U. and Bryant, D. A. "Molecular Contacts for Chlorosome Envelope Proteins Revealed by Cross-Linking Studies with Chlorosomes from Chlorobium tepidum". "Biochemistry". 2006. Volume 45. p. 9095 -9103 [6]
[8] http://en.wikipedia.org/wiki/Fenna-Matthews-Olson_complex
[9] http://en.wikipedia.org/wiki/Reaction_center
Acknowledgements
The authors would like to thank Dr. D. A. Bryant and A. M. Garcia (The Pennsylvania State University) for sharing the most recent information about genome sequencing of Cab. thermophilum and their insightful suggestions about this webpage. We are thankful to Dr. D. A. Bryant for providing the copyright permission for figures from his publications. We would like to thank Dr J. Lennon, Dr. N. Walker, Dr. F. B. Dazzo, and Dr. R. Lenski at Michigan State University for including microbeWIKI project in Microbial Ecology (MMG-425) course curriculum. Farhan Ahmad would like to thank Dr. Syed A. Hashsham (advisor), for providing Ph.D. oppourtunity in the Environmental Genomics Lab [7]
Author
Page authored by Ahmad Farhan and Anadumaka V. Chike, student of Prof. Jay Lennon at Michigan State University.