Clostridium baratii: Difference between revisions
No edit summary |
No edit summary |
||
| (34 intermediate revisions by the same user not shown) | |||
| Line 9: | Line 9: | ||
===Higher order taxa=== | ===Higher order taxa=== | ||
Bacteria; Firmicutes; Clostridia; Clostridiales; | Bacteria; Firmicutes; Clostridia; Clostridiales; Clostridiaceae; Clostridium | ||
===Species=== | ===Species=== | ||
''Clostridium baratii'' | |||
{| | {| | ||
| height="10" bgcolor="#FFDF95" | | | height="10" bgcolor="#FFDF95" | | ||
'''Clostridium | '''''Clostridium baratii'' NCBI: [https://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?mode=Info&id=1561&lvl=3&lin=f&keep=1&srchmode=1&unlock Taxonomy]''' | ||
|} | |} | ||
==Description and significance== | ==Description and significance== | ||
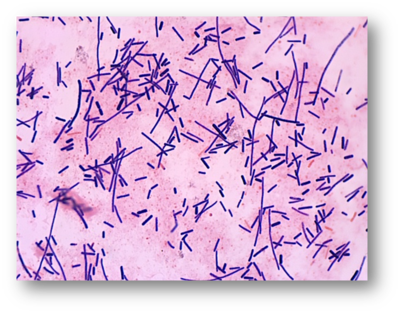
''Clostridium baratii'' derives its name from the French bacteriologist Barat. This species of bacteria was first identified in 1938 by Prévot. ''C. baratii'' are gram positive rod-shaped bacteria, the colonies of which can appear round or irregular and have a diameter of 2mm or smaller. ''C. baratii'' do not typically appear in groups and are nonmotile. This species can produce spores which are round or oval and located at or near the terminus of the cell(1). | ''Clostridium baratii'', a member of the genus [[https://microbewiki.kenyon.edu/index.php/Clostridium Clostridium]], derives its name from the French bacteriologist Barat (1). This species of bacteria was first identified in 1938 by Prévot, though little is recorded about this first isolation (1). ''C. baratii'' are gram positive, rod-shaped bacteria, the colonies of which can appear round or irregular and have a diameter of 2mm or smaller (1). ''C. baratii'' do not typically appear in groups and are nonmotile. This species can produce spores which are round or oval and located at or near the terminus of the cell (1). | ||
''C. baratii'' produces toxins which cause botulism, most notably in infants (2, 3). Infant botulism is the most common type of botulism diagnosed today, making the study of ''C. baratii'' and its pathogenicity of particular interest (2). | |||
==Genome and genetics== | ==Genome and genetics== | ||
The genus ''Clostridium'' is part of the prokaryotic branch ''Firmicutes'' (1). ''C. baratii'' is part of Cluster I of the ''Clostridium' genus and is most closely related to the species '' | The genus ''Clostridium'' is part of the prokaryotic branch ''Firmicutes'' (1). ''C. baratii'' is part of Cluster I of the ''Clostridium'' genus and is most closely related to the species ''C. sardiniense'' according to the phylogenic tree (Collin et al., as cited in 1). ''C. baratii'' is also closely related to several species of ''Eubacteria'', particularly ''Eubacterium budayi'', with which it shares 99.5 percent of its genome according to 16S rRNA genetic sequencing (1). | ||
The genome of ''C. baratii'' shows 28 percent G+C content (1) and the genome of ''C. baratii'' strain XCM is 3083960 base pairs in length (4). This strain of ''C. baratii'' has 3027 genes, 2920 of which code for various proteins (4). | The genome of ''C. baratii'' shows 28 percent G+C content (1) and the genome of ''C. baratii'' strain XCM is 3083960 base pairs in length (4). This strain of ''C. baratii'' has 3027 genes, 2920 of which code for various proteins (4). The full genome sequence for this strain is not currently available online. | ||
The complete genome sequencing of ''C. baratii'' strain Sullivan can be found on the [[https://www.ncbi.nlm.nih.gov/nuccore/NZ_CP006905.1 National Center for Biotechnology Information database]]. This strain of ''C. baratii'' is 3153266 base pairs in length and contains 3210 genes (9). This genome was sequenced using 454 and Illumina technologies (9). | |||
==Nutrition and metabolism== | ==Nutrition and metabolism== | ||
''C. baratii'' is an anaerobic bacteria species (1). Through fermentation of glucose in | ''C. baratii'' is an anaerobic bacteria species (1). Through fermentation of glucose in Peptone Yeast Glucose broth, ''C. baratii'' can produce lactic acid, acetic acid, and butyric acid, as well as the potentially volatile product hydrogen gas (1). This bacteria also produces ammonia in deep agar cultures (1). Additionally, ''C. baratii'' converts pyruvate to acetate and butyrate but does not utilize lactate (1). | ||
''C. baratii'' is grown in the laboratory in Peptone Yeast Glucose broth and on blood agar plates and produces the most abundant growth at temperatures between 30 and 35 degrees Celsius (1). It requires salinity below 6.5 percent sodium chloride (1). ''C. baratii'' grows in PYG broth, which has a nearly neutral pH, but over several days of culture growth the pH lowers to between 4.5 and 4.8 (1, catalog). | ''C. baratii'' is grown in the laboratory in Peptone Yeast Glucose broth and on blood agar plates and produces the most abundant growth at temperatures between 30 and 35 degrees Celsius (1). It requires salinity below 6.5 percent sodium chloride (1). ''C. baratii'' grows in PYG broth, which has a nearly neutral pH, but over several days of culture growth the pH lowers to between 4.5 and 4.8 (1, catalog). | ||
| Line 45: | Line 44: | ||
==Pathology== | ==Pathology== | ||
C. baratii produces neurotoxins which cause botulism in humans (5). Botulism is most commonly caused by neurotoxins produced by C. botulinum, but C. baratii produces similar neurotoxins (5, 6). These toxins have been designated by letters, and human botulism has been caused by toxins A, B, E, and F (6). The neurotoxins which cause botulism are the most potent toxins currently known and cause paralysis through the inhibition of acetlylcholine release in human nerve endings (5). Symptoms may begin as a headache and vomiting and progress to proximal and then distal paralysis (6). Eventually respiratory failure may occur due to paralysis and the morality rate for botulism is currently only 3 to 5 percent, part due to mechanical respiration technology (5). | ''C. baratii'' produces neurotoxins which cause botulism in humans (5). Botulism is most commonly caused by neurotoxins produced by ''C. botulinum'', but ''C. baratii'' produces similar neurotoxins (5, 6). These toxins have been designated by letters, and human botulism has been caused by toxins A, B, E, and F (6). The neurotoxins which cause botulism are the most potent toxins currently known and cause paralysis through the inhibition of acetlylcholine release in human nerve endings (5). Symptoms may begin as a headache and vomiting and progress to proximal and then distal paralysis (6). Eventually respiratory failure may occur due to paralysis and the morality rate for botulism is currently only 3 to 5 percent, in part due to mechanical respiration technology (5). | ||
A case of botulism is confirmed when the presence of botulism-causing neurotoxins in a patient’s vomit or stool is identified by a laboratory using either mouse bioassays or enzyme-linked immunosorbent assays (5). While there is no vaccine for botulism, antitoxin is currently available to treat patients diagnosed with botulism, and produces best results when administered within 24 hours of the onset of symptoms, as it may only bind with free neurotoxins and cannot remove those that have already affected neuromuscular junctions. (5). | A case of botulism is confirmed when the presence of botulism-causing neurotoxins in a patient’s vomit or stool is identified by a laboratory using either mouse bioassays or enzyme-linked immunosorbent assays (5). While there is no vaccine for botulism, antitoxin is currently available to treat patients diagnosed with botulism, and produces best results when administered within 24 hours of the onset of symptoms, as it may only bind with free neurotoxins and cannot remove those that have already affected neuromuscular junctions. (5). | ||
==Current Research== | ==Current Research== | ||
Recently, researchers developed PCR tests which would allow strains of '' | Recently, researchers developed PCR tests which would allow strains of ''C. baratii'' and ''C. botulinum'' which produce botulism-causing neurotoxins A, B, E, and F to be identified much more rapidly than the typically used method of mouse bioassays (7). The practical clinical benefits of being able to identify these bacteria more readily are numerous, and the results of the study were promising as they tested the validity of the PCR tests using an infected sample of meat (7). The researchers began by sequencing two genes on each strain of bacteria: the BoNT (botulism neurotoxin) gene corresponding to a specific toxin type in each strain, and the NTNH (nontoxin-nonhaemagglutinin) gene, which is associated with and located upstream of the BoNT gene (7). Using these results, the researchers developed one universal primer pair and probe, four toxin type-specific primer pairs and probes, and one NTNH primer pair and probe to identify the correct genes in each strand of bacteria as part of a PCR test (7). The researchers tested the effectiveness of their work by using cultures of ''C. baratii'' and ''C. botulinum'' strains in PCR tests; the tests successfully identified the toxin-producing genes (BoNT/F in ''C. baratii'') (7). Nontoxic strains of ''Clostridia'' tested negative, indicating successful development of PCR tests that adequately identify specific BoNT-producing strains of bacteria (7). | ||
Recently, the first French cases of botulism in which ''C. baratii'' was found to be the cause were reported (8). Citing additional studies, the researchers note that occurrences of botulism resulting from ''C. baratii'' toxin Type F are rare and have generally been reported in the US alone, making these cases in France highly unique (8). The reported cases were two women, a woman in her sixties and a woman in her twenties; the two were relatives who had shared meals in the weeks preceding their illness (8). The women’s symptoms included respiratory distress and paralysis, and both women required respiratory assistance, with one patient hospitalized for 46 days and the other for 19 days (8). ''C. baratii'' was found in the stool of both patients, and botulism neurotoxin Type F was found in the serum of both patients (higher levels of toxin were found in the patient with more severe symptoms) (8). Investigators isolated one family meal as the likely source of infection, but no toxins were ever isolated from the food samples which remained, and no foods typically associated with botulism were consumed at the meal (8). Additionally, no other family members became ill (8). As these two cases were the first of their kind in France, there is still much to be learned about the presence and pathogenicity of ''C. baratii'' in France. | |||
==References== | ==References== | ||
[1] De Vos P, Garrity GM, Jones D, Krieg NR, Ludwig W, Rainey FA, Schleifer KH, Whitman WB, editors. Bergey’s manual of systematic bacteriology (2nd ed., vol 3). Dordrecht: Springer; 2009. | [1] De Vos P, Garrity GM, Jones D, Krieg NR, Ludwig W, Rainey FA, Schleifer KH, Whitman WB, editors. Bergey’s manual of systematic bacteriology (2nd ed., vol 3). Dordrecht: Springer;2009:756-757. | ||
[https://www.ncbi.nlm.nih.gov/pubmed/26593441 [2]] Shirey TB, Dykes JK, Lúquez C, Maslanka SE, Raphael BH. Characterizing the fecal microbiota of infants with botulism. Microbiome. 2015;3(54):1-13. | |||
[http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=21360 [3]] Tréhard H, Poujol I, Mazuet C, Blanc Q, Gillet Y, Rossignol F, Popoff M, Jourdan Da Silva N. A cluster of three cases of botulism due to ''Clostridium baratii'' Type F, France, August 2015. Eurosurveillance. 2016;21(4):2-5 | |||
[ | [https://img.jgi.doe.gov/cgi-bin/m/main.cgi?section=TaxonDetail&page=taxonDetail&taxon_oid=2648501875#browse [4]] Joint Genome Institute. Clostridium baratii XCM. 2016. | ||
[ | [https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3148622/ [5]] Thavaselvam D, Vijayaraghavan R. Biological warfare agents. Journal of Pharmacy and Bioallied Sciences. 2010;2(3):n.p. | ||
[ | [http://ecdc.europa.eu/en/publications/Publications/botulism-scientific-advice-type-F-botulism.pdf [6]] European Centre for Disease Prevention and Control. Technical Report: Scientific Advice on Type F Botulism. 2013:1-13. | ||
[ | [http://onlinelibrary.wiley.com/doi/10.1111/j.1365-2672.2009.04215.x/abstract;jsessionid=55B2E9635DB905FD95AC84EA2ED46C5F.f03t02 [7]] Fach P, Micheau P, Mazuet C, Perelle S, Popoff M. Development of real-time PCR tests for detecting botulinum neurotoxins A, B, E, F producing ''Clostridium botulinum, Clostridium baratii'' and ''Clostridium butyricum''. Journal of Applied Microbiology. 2009;107(2):465-473. | ||
[ | [http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=21031 [8]] Castor C, Mazuet C, Saint-Leger M, Vygen S, Coutureau J, Durand M, Popoff MR, Jourdan Da Silva N. Cluster of two cases of botulism due to ''Clostridium baratii'' Type F in France, November 2014. Eurosurveillance. 2015;20(6):1-3. | ||
[ | [https://www.ncbi.nlm.nih.gov/nuccore/NZ_CP006905.1 [9]] Smith TJ, Hill KK, Xie G, Foley BT, Williamson CH, Foster JT, Johnson SL, Chertkov O, Teshima H, Gibbons HS, Johnsky LA, Karavis MA, Smith LA. Clostridium baratii str. Sullivan, complete genome. Infect. Genet. Evol. 2015;30:102-113. | ||
Latest revision as of 04:51, 3 May 2017

Classification
Higher order taxa
Bacteria; Firmicutes; Clostridia; Clostridiales; Clostridiaceae; Clostridium
Species
Clostridium baratii
|
Clostridium baratii NCBI: Taxonomy |
Description and significance
Clostridium baratii, a member of the genus [Clostridium], derives its name from the French bacteriologist Barat (1). This species of bacteria was first identified in 1938 by Prévot, though little is recorded about this first isolation (1). C. baratii are gram positive, rod-shaped bacteria, the colonies of which can appear round or irregular and have a diameter of 2mm or smaller (1). C. baratii do not typically appear in groups and are nonmotile. This species can produce spores which are round or oval and located at or near the terminus of the cell (1).
C. baratii produces toxins which cause botulism, most notably in infants (2, 3). Infant botulism is the most common type of botulism diagnosed today, making the study of C. baratii and its pathogenicity of particular interest (2).
Genome and genetics
The genus Clostridium is part of the prokaryotic branch Firmicutes (1). C. baratii is part of Cluster I of the Clostridium genus and is most closely related to the species C. sardiniense according to the phylogenic tree (Collin et al., as cited in 1). C. baratii is also closely related to several species of Eubacteria, particularly Eubacterium budayi, with which it shares 99.5 percent of its genome according to 16S rRNA genetic sequencing (1).
The genome of C. baratii shows 28 percent G+C content (1) and the genome of C. baratii strain XCM is 3083960 base pairs in length (4). This strain of C. baratii has 3027 genes, 2920 of which code for various proteins (4). The full genome sequence for this strain is not currently available online.
The complete genome sequencing of C. baratii strain Sullivan can be found on the [National Center for Biotechnology Information database]. This strain of C. baratii is 3153266 base pairs in length and contains 3210 genes (9). This genome was sequenced using 454 and Illumina technologies (9).
Nutrition and metabolism
C. baratii is an anaerobic bacteria species (1). Through fermentation of glucose in Peptone Yeast Glucose broth, C. baratii can produce lactic acid, acetic acid, and butyric acid, as well as the potentially volatile product hydrogen gas (1). This bacteria also produces ammonia in deep agar cultures (1). Additionally, C. baratii converts pyruvate to acetate and butyrate but does not utilize lactate (1).
C. baratii is grown in the laboratory in Peptone Yeast Glucose broth and on blood agar plates and produces the most abundant growth at temperatures between 30 and 35 degrees Celsius (1). It requires salinity below 6.5 percent sodium chloride (1). C. baratii grows in PYG broth, which has a nearly neutral pH, but over several days of culture growth the pH lowers to between 4.5 and 4.8 (1, catalog).
C. baratii reduces the indicator dyes neutral red and resazurin (1).
Pathology
C. baratii produces neurotoxins which cause botulism in humans (5). Botulism is most commonly caused by neurotoxins produced by C. botulinum, but C. baratii produces similar neurotoxins (5, 6). These toxins have been designated by letters, and human botulism has been caused by toxins A, B, E, and F (6). The neurotoxins which cause botulism are the most potent toxins currently known and cause paralysis through the inhibition of acetlylcholine release in human nerve endings (5). Symptoms may begin as a headache and vomiting and progress to proximal and then distal paralysis (6). Eventually respiratory failure may occur due to paralysis and the morality rate for botulism is currently only 3 to 5 percent, in part due to mechanical respiration technology (5).
A case of botulism is confirmed when the presence of botulism-causing neurotoxins in a patient’s vomit or stool is identified by a laboratory using either mouse bioassays or enzyme-linked immunosorbent assays (5). While there is no vaccine for botulism, antitoxin is currently available to treat patients diagnosed with botulism, and produces best results when administered within 24 hours of the onset of symptoms, as it may only bind with free neurotoxins and cannot remove those that have already affected neuromuscular junctions. (5).
Current Research
Recently, researchers developed PCR tests which would allow strains of C. baratii and C. botulinum which produce botulism-causing neurotoxins A, B, E, and F to be identified much more rapidly than the typically used method of mouse bioassays (7). The practical clinical benefits of being able to identify these bacteria more readily are numerous, and the results of the study were promising as they tested the validity of the PCR tests using an infected sample of meat (7). The researchers began by sequencing two genes on each strain of bacteria: the BoNT (botulism neurotoxin) gene corresponding to a specific toxin type in each strain, and the NTNH (nontoxin-nonhaemagglutinin) gene, which is associated with and located upstream of the BoNT gene (7). Using these results, the researchers developed one universal primer pair and probe, four toxin type-specific primer pairs and probes, and one NTNH primer pair and probe to identify the correct genes in each strand of bacteria as part of a PCR test (7). The researchers tested the effectiveness of their work by using cultures of C. baratii and C. botulinum strains in PCR tests; the tests successfully identified the toxin-producing genes (BoNT/F in C. baratii) (7). Nontoxic strains of Clostridia tested negative, indicating successful development of PCR tests that adequately identify specific BoNT-producing strains of bacteria (7).
Recently, the first French cases of botulism in which C. baratii was found to be the cause were reported (8). Citing additional studies, the researchers note that occurrences of botulism resulting from C. baratii toxin Type F are rare and have generally been reported in the US alone, making these cases in France highly unique (8). The reported cases were two women, a woman in her sixties and a woman in her twenties; the two were relatives who had shared meals in the weeks preceding their illness (8). The women’s symptoms included respiratory distress and paralysis, and both women required respiratory assistance, with one patient hospitalized for 46 days and the other for 19 days (8). C. baratii was found in the stool of both patients, and botulism neurotoxin Type F was found in the serum of both patients (higher levels of toxin were found in the patient with more severe symptoms) (8). Investigators isolated one family meal as the likely source of infection, but no toxins were ever isolated from the food samples which remained, and no foods typically associated with botulism were consumed at the meal (8). Additionally, no other family members became ill (8). As these two cases were the first of their kind in France, there is still much to be learned about the presence and pathogenicity of C. baratii in France.
References
[1] De Vos P, Garrity GM, Jones D, Krieg NR, Ludwig W, Rainey FA, Schleifer KH, Whitman WB, editors. Bergey’s manual of systematic bacteriology (2nd ed., vol 3). Dordrecht: Springer;2009:756-757.
[2] Shirey TB, Dykes JK, Lúquez C, Maslanka SE, Raphael BH. Characterizing the fecal microbiota of infants with botulism. Microbiome. 2015;3(54):1-13.
[3] Tréhard H, Poujol I, Mazuet C, Blanc Q, Gillet Y, Rossignol F, Popoff M, Jourdan Da Silva N. A cluster of three cases of botulism due to Clostridium baratii Type F, France, August 2015. Eurosurveillance. 2016;21(4):2-5
[4] Joint Genome Institute. Clostridium baratii XCM. 2016.
[5] Thavaselvam D, Vijayaraghavan R. Biological warfare agents. Journal of Pharmacy and Bioallied Sciences. 2010;2(3):n.p.
[6] European Centre for Disease Prevention and Control. Technical Report: Scientific Advice on Type F Botulism. 2013:1-13.
[7] Fach P, Micheau P, Mazuet C, Perelle S, Popoff M. Development of real-time PCR tests for detecting botulinum neurotoxins A, B, E, F producing Clostridium botulinum, Clostridium baratii and Clostridium butyricum. Journal of Applied Microbiology. 2009;107(2):465-473.
[8] Castor C, Mazuet C, Saint-Leger M, Vygen S, Coutureau J, Durand M, Popoff MR, Jourdan Da Silva N. Cluster of two cases of botulism due to Clostridium baratii Type F in France, November 2014. Eurosurveillance. 2015;20(6):1-3.
[9] Smith TJ, Hill KK, Xie G, Foley BT, Williamson CH, Foster JT, Johnson SL, Chertkov O, Teshima H, Gibbons HS, Johnsky LA, Karavis MA, Smith LA. Clostridium baratii str. Sullivan, complete genome. Infect. Genet. Evol. 2015;30:102-113.
Authored by Emma Wingert, a student of CJ Funk at John Brown University
