Commercial applications of Rhodopseudomonas palustris
Introduction
Rhodopseudomonas palustris is an Alphaproteobacteria under the genus Rhodopseudomonas. R. palustris is a non-sulfur purple bacterium that is gram-negative with a rod shape. Cells are motile and the organism reproduces by means of budding. R. palustris is found in both aerobic and anaerobic environments including a wide variety of marine and soil ecosystems; coastal marine sediment and waste water treatment facilities for example [1]. Typically non-sulfur purple bacteria are phototrophic, however R. palustris has the capability to switch between different forms of metabolism depending on environmental conditions: photoautotrophy, photoheterotrophy, chemoautotrophy, and chemoheterotrophy.
Rhodopseudomonas palustris is the focus of extensive research because it has a multitude of potential metabolic processes. Research focusing on bioremediation and removal of waste is interested in R. palustris for its potential to break down aromatic compounds and waste in polluted environments. Rhodopseudomonas also has the capability to produce hydrogen as a product of nitrogen fixation, which researchers are looking into as a form of alternate energy produced from biofuel. R. palustris has also displayed the capability to generate electricity further increasing the scope of research surrounding the microbe. Climate change induced by global warming and pollution of environments are some of the largest environmental problems facing the modern world. R. palustris is interesting in that it could hold the potential to solving not just one, but both of these serious environmental issues. The extensive research on this microbe presents a possibility for unique commercial applications of the microbe as a source of alternate energy and in waste management and removal.
Metabolism
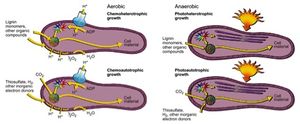
R. palustris has the capability to metabolize under aerobic and anaerobic conditions, meaning the organism can produce energy with or without oxygen present. Anaerobically, R. palustris can utilize photoheterotrophy and photoautotrophy, or photosynthesis[2]. The cell has the capability to fix carbon dioxide as biomass through photosynthesis, similar to green plants, but also R. palustris has the capability to obtain energy from light and other organic carbon forms via photoheterotrophy[3]. Under photoheterotrophic conditions, R. palustris utilizes organic compounds, most notably lignin monomers as a source of carbon[4]. When the cell is in the presence of conditions conducive to phototrophy, lamellar intracytoplasmic membranes hold bacteriochlorophylls and carotenoids, the photosynthetic pigments of the cell that make up the light harvesting complex, next to the cell membrane, which allows sunlight to be absorbed by the photosynthetic pigments[5]. The light energy absorbed is used to move electrons eventually resulting in the production of adenosine triphosphate (ATP). ATP is used as energy within the cell while the fixation of carbon dioxide or other organic compounds allows for the production of biomass and new cell material. Research has found that, R. palustris is able to respond to varying levels of light in its environment; in environments with lower light intensity the cell increases the amount of light harvesting complexes present to more effectively capture light energy[6]. Like photosynthetic plants, R. palustris possesses genes, which encode for RuBisCo, the enzyme responsible for the fixation of carbon dioxide in plants[7]. RuBisCo is used for the same fixation function in R. palustris photosynthesis, however unlike in plant photosynthesis oxygen is not a product of photosynthesis, as water is not split in the reaction and R. palustris utilizes thiosulfate, hydrogen and other inorganic electron donors[8].
Research has shown that R. palustris has genes that resemble genes found in oxygenic photosynthetic bacteria that regulate a form of circadian clock for the organism[9]. While photosynthesis generates large amounts of energy for the cell during light hours, the energy produced would be limited during dark hours. The presence of these circadian clock genes would suggest that there is circadian regulation of reactions, in the cell notably for the process of nitrogen fixation[10]. Rhodopseudomonas is an example of a microbe that performs anoxic photosynthesis in which oxygen is not generated and instead nitrogen gas is fixed. R. palustris possesses genes that encode nitrogenase enzymes, which allows the organisms to take in atmospheric nitrogen gas and fix the nitrogen into ammonia and other fixed nitrogen forms.
When oxygen is present in the environment, R. palustris can utilize forms of chemotrophy for metabolizing energy in the cell. These processes are regulated by the abundance of compounds present in the environment, as R. palustris has the flexibility to metabolize as a chemoautotroph or as a chemoheterotroph in the presence of oxygen[11]. As a chemoautotroph, R. palustris uses inorganic organic electron donors, like thiosulfate and hydrogen, to synthesis organic compounds using carbon dioxide as an oxidizer of the inorganic electron donor[12]. If carbon dioxide is not present in the system, or is present in small amounts, R. palustris can synthesize other organic compounds, namely lignin, in order for the cell to form its own organic compounds for use in the cell. Both of these metabolic forms in R. palustris utilize proton motive force, a hydrogen ion gradient in and around the cell, to transform adenosine diphosphate into adenosine triphosphate to provide the energy required within the cell. Furthermore, both chemotrophic metabolic processes result in the production of water, since oxygen serves as the final electron acceptor of the system. Research has shown however that R. palustris can utilize dimethyl sulfoxide, potassium nitrate, or sodium nitrite as terminal electron acceptors instead of oxygen[13].
Biodegredation
The function of most importance in studying R. palustris capabilities as a biodegradation tool is its ability to breakdown aromatic compounds. Aromatic structures, on a most basic level defined by having a planar ring, are commonly found in many, if not most, of the pollutants that can be harmful to environmental systems. Industrial wastes, sewage, plastics, and pesticides used in agriculture are all common pollutants and all contain aromatic structures of some form. One of the most common forms of aromatic pollutants is benzene, which is found in many commercial and waste products. The danger of benzene and many other aromatics is the ease of solubility, making it easy for these pollutants to enter water systems and cause harm[14]. These aromatic pollutants pose a serious threat to the health of many ecosystems and can also have adverse health consequences when they come into human contact.
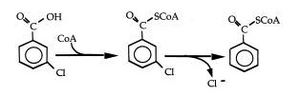
The ability of R. palustris to degrade aromatic compounds has been researched extensively. This research has lead to a fairly complete understanding of the mediated pathway used to breakdown 3-chlorobenzoate, a common aromatic form observed in many pollutants. R. palustris utilizes a reductive coenzyme to convert, via reduction and dehalogentation, 3-chlorobenzoate into acetyl CoA and carbon dioxide[15]. In chemotrophic metabolism, the cell utilizes acetyl CoA as an energy source, incorporating the newly formed acetyl CoA compound into the tricarboxylic acid cycle. This process is unique in R. palustris in that the aromatic compound, in the pathway of 3-chlorobenzoate degradation, is the substrate for complete dehalogenation[16]. Further research has shown that R. palustris has been able to reductively dehalogenated and assimilate many different forms of aromatic acids, phenolic, dihydroxylated, and methoxylated, as well as aromatic aldehydes, and hydroaromatic acids as R. palustris growth has been observed on these substrates with and without the presence of oxygen[17]. R. palustris has also demonstrated the ability to breakdown halocarboxylic acids; another common environmental pollutant found in aquatic and moist soil environments[18].
R. palustris degradation of aromatics is unique not just because of the diversity of the aromatics it can breakdown, but R. palustris is also unique in the multitude of metabolic processes it can utilize to break down the aromatic compounds. In fact research has shown that even strains of R. palustris with similar substrate utilization patterns can have differences in degredation[19]. Furthermore, research indicates that R. palustris can utilize its different metabolic pathways, aerobic and anaerobic, to mediate the breakdown of a wide range of aromatic compounds[20]. The ability to breakdown these pollutant aromatic structures is important to understand commercially, as this capability of R. palustris could prove to be harnessed and utilized in order to remove a wide range of pollutants from many different environments. The diversity of aromatics that can be degraded allows for wide coverage of possible pollutants R. palustris can break down. The ability to utilize different metabolic pathways to degrade the aromatics allows the microbe to remove pollutants in a wide range of environments as well. Research has already been conducted indicating the success of R. palustris removing odorous pollutants from wastewater lagoons[21] and eutrophic ponds[22].
Electricity Generation
Some bacteria have the ability to generate electricity in microbial fuel cells (MFCs), which is a bioelectrical system that produces current by using bacteria, although typically power production is very low. However, one stain of R. palustris, Rhodopseudomonas palustris DX-1, has been shown to produce higher power output (2720 mW/m2) when isolated than mixed cultures produced[23]; R. palustris is the first Alphaproteobacteria known to produce high power densities in this fashion. Research has demonstrated that DX-1 has the capability to generate power from a wide range of organic matter using a variety of different substrates like volatile acids and thiosulfate, which is accomplished by utilizing the organism’s different modes of metabolism. While the power generated by the microbe R. palustris DX-1 will not solve an impending energy crisis, this discovery is highly useful in further analysis of power generation in microbial fuel cells.
Another strain of R. palustris has been recorded to have electricity producing potential as well, the TIE-1 strain. Rhodopseudomonas palustris TIE-1 performs extracellular electron transfer, using solid material and minerals as electron acceptors or donors. Without the presence of oxygen, the TIE-1 strain metabolizes as a photoautotroph utilizing most notably Fe (II) as an electron donor among other inorganic compounds[24]. The TIE-1 strain also metabolizes as a photohetrotroph if organic compounds are available and as a chemoheterotroph in the dark. TIE-1 takes in electrons from iron and other minerals and uses carbon dioxide as an electron acceptor in order to convert the electrons into energy utilizing the RuBisCo enzyme[25]. While RuBisCo expression is stimulated by light, electron uptake can also occur in the dark, using the pioABC operon, which encodes a protein system responsible for iron oxidation[26]. Effectively, TIE-1 creates a charge by utilizing mineral in the soil and also utilizing light energy from the surface.
DX-1 and TIE-1 are important discoveries and create further areas of research regarding R. palustris. With current research and technology however, harnessing electrical capabilities of R. palustris is not feasible as DX-1 does not produce enough electricity to self sustain wastewater treatment as analyzed in research[27]. TIE-1 serves as a prospect in the manufacturing of batteries, however this technology and research has not been fully realized[28].
Hydrogen Production
A growing global concern is the world’s reliance on fossil fuels as a source of energy. Countless hours of research and large monetary investments have been made to create and harness alternate forms of energy. Biofuels are a burgeoning area of study within the alternate energy field; the focus being to find organismal biological processes which create, in some form, a viable source of energy that can then be captured and used for human use. R. palustris is an interesting organism analyzed in the study of biofuels as it produces hydrogen as a result of fixing nitrogen. This byproduct hydrogen produced could potentially be tapped into as an energy source for humans, resulting in a large investment of labor into analyzing R. palustris ability to produce viable biofuel. Hydrogen can be a useful biofuel because it represents a much cleaner energy source than fossil fuels; combustion of hydrogen only produces water, unlike fossil fuel combustion that releases carbon dioxide and other greenhouse gases which serve only to worsen global warming and climate change.
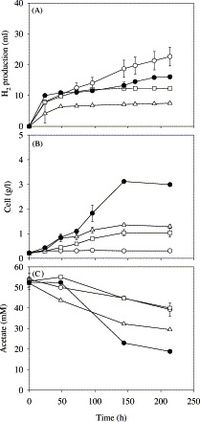
Other purple non-sulfuric bacteria perform nitrogen fixation, however R. palustris is unique in that its genome encodes for three different nitrogen fixing enzymes[29]. The most important to R. palustris potential as a hydrogen producer of the three nitrogenases is the vanadium-containing nitrogenase. The vanadium-containing nitrogenase produces three times more hydrogen than nitrogenases of other nitrogen-fixing bacteria[30]. The ability of R. palustris to produce significantly larger amounts of hydrogen than other nitrogen-fixing bacteria is what lends credit to R. palustris’ ability to generate hydrogen as a form of alternate energy. An isolated strain of R. palustris, Rhodopseudomonas palustris P4 is able to utilize a wide range of organic compounds including monosaccharaides, disaccharides, and starches to produce hydrogen from fermentation[31]. P4 is also able to produce hydrogen from carbon monoxide via a water-gas shift reaction in addition to the previously described anaerobic respiration[32]. Research has shown that fermentation production by the P4 strain is much faster, but produces significantly less hydrogen than the gas-shift reaction or nitrogen fixation[33]. However, the P4 strain is unique in that it can preform fermentation under dark circumstances, but also in light environments; the combined dark and light fermentation process increases the hydrogen production by two-fold compared to a less complex strict dark fermentation process[34].
Other research has also aimed to increase hydrogen production yield. Some studies have coupled R. palustris with other forms of nitrogen-bacteria, cyanobacteria. Cyanobacteria possess hydrogenases, which consume the hydrogen produced as a result of nitrogen fixation, however mutant strains can be engineered with no hydrogenases. The coupling of R. palustris with these mutant cyanobacteria only serve to increase hydrogen output, a 4.8 fold increase in hydrogen production as observed in the research[35]. While more research is required and advances are necessary in modern technologies for R. palustris hydrogen production to become a viable source of energy, the prospect of one day fueling your car or lighting your house with biofuel energy produced by R. palustris is not as farfetched an idea as it would have been decades ago.
Conclusion
Rhodopseudomonas palustris is a microbe that has sparked intense research across the globe due to the potentially profound results that may be found. R. palustris has the extreme potential to change the way humans live on this planet, most effectively by providing an alternate fuel source to fossil fuels. The environmental ramifications of R. palustris research are astounding as this microbe has the capability to change how the world addresses issues of pollution, energy production, and possibly even electricity generation. Rhodopseudomonas is able to have this astounding potential because of its unique ability to switch between four main sources of metabolism depending on what is available to the organism in the environment. R. palustris can metabolize in a large variance of environments; aerobic or anaerobic, euphotic or aphotic, organic materials available or inorganic materials available. The ability to circumvent between different forms of metabolism is what affords R. palustris’ reputation and potential within the biotechnology field.
R. palustris has the potential to degrade numerous aromatic structures; structures common in many of the worst environmental pollutants. Agricultural pesticides that run off, petroleum based plastics, oil, and sewage all pollute and threaten aquatic ecosystems across the globe, but R. palustris has the ability to break down all of these toxins and pollutants. This is an incredibly valuable asset commercially as R. palustris could in the future be employed to clean up polluted sites and environments as it has already been employed in wastewater treatment facilities. R. palustris has the potential ability, if it can be harnessed, to clean up anthropogenic pollution that has degraded countless environments in a more effective way that has ever been achieved in the environmental sector before.
Even further, R. palustris also has a large amount of intrigue as a potential replacement to fossil fuels. The energy production capabilities of R. palustris, if research and technology can make it feasible to capture and harness such energy, could radically change the way in which the world operates in the future. Alternative energy sources must be found and utilized if the affects of global warming and climate change are to be quelled and, if even possible, reversed. The production of clean burning hydrogen fuel created by R. palustris may just be the answer the alternative energy sector has been searching for in trying to find a feasible and efficient replacement to fossil fuels. Furthermore, while the affects may be less profound or widespread, R. palustris also presents potential in better understanding microbial electricity production. Strains of R. palustris have been shown to produce electricity; the DX-1 strain produces at a higher power density than was expected in MFC[36]. The commercial benefits of these electricity producing strains, while not world changing like providing an alternate energy source, consist of providing better insight into microbial electricity generation, self-sustaining wastewater treatment facilities[37], and even battery production[38].
Rhodopseudomonas palustris is a unique microbe that has the potential to provide multiple commercial applications. It will surely be interesting to follow the continued research of R. palustris and hopefully the implementation of future technologies based on further understanding of this microbe.
References
- ↑ Larimer F. et al. “Complete genome sequence of the metabolically versatile photosynthetic bacterium Rhodopseudomonas palustris”. Nature Biotechnology. 2004. Volume 22, p.55-61
- ↑ Larimer F. et al. “Complete genome sequence of the metabolically versatile photosynthetic bacterium Rhodopseudomonas palustris”. Nature Biotechnology. 2004. Volume 22, p.55-61
- ↑ Larimer F. et al. “Complete genome sequence of the metabolically versatile photosynthetic bacterium Rhodopseudomonas palustris”. Nature Biotechnology. 2004. Volume 22, p.55-61
- ↑ Larimer F. et al. “Complete genome sequence of the metabolically versatile photosynthetic bacterium Rhodopseudomonas palustris”. Nature Biotechnology. 2004. Volume 22, p.55-61
- ↑ Mehrabi,S. et al." Identification and characterization of Rhodopseudomonas spp., a purple, non-sulfur bacterium from microbial mats"."Biomolecular Engineering". 2001.Volume 18,p.49-56.
- ↑ Larimer F. et al. “Complete genome sequence of the metabolically versatile photosynthetic bacterium Rhodopseudomonas palustris”. Nature Biotechnology. 2004. Volume 22, p.55-61
- ↑ Scheuring, S. “The Photosynthetic Apparatus of Rhodopseudomonas palustris: Structures and Organization”. Journal of Molecular Biology. 2006.Volume 358, Issue 1, p.83-96.
- ↑ Scheuring, S. “The Photosynthetic Apparatus of Rhodopseudomonas palustris: Structures and Organization”. Journal of Molecular Biology. 2006.Volume 358, Issue 1, p.83-96.
- ↑ Scheuring, S. “The Photosynthetic Apparatus of Rhodopseudomonas palustris: Structures and Organization”. Journal of Molecular Biology. 2006.Volume 358, Issue 1, p.83-96.
- ↑ Scheuring, S. “The Photosynthetic Apparatus of Rhodopseudomonas palustris: Structures and Organization”. Journal of Molecular Biology. 2006.Volume 358, Issue 1, p.83-96.
- ↑ Egland, P. et al. “Reductive Coenzyme A-Mediated Pathway for 3-Chlorobenzoate Degradation in the Phototrophic Bacterium Rhodopseudomonas palustris”. Applied and Environmental Microbiology. 2001. Volume 67, Issue 3, p. 1396-1399.
- ↑ Larimer F. et al. “Complete genome sequence of the metabolically versatile photosynthetic bacterium Rhodopseudomonas palustris”. Nature Biotechnology. 2004. Volume 22, p.55-61
- ↑ Lang, F., Oesterhelt, D. "Microaerophilic growth and induction of the photosynthetic reaction center in Rhodopseudomonas viridis." Journal of Bacteriology, 1989. Volume 171, Issue 5. American Society for Microbiology. (2327-2834).
- ↑ Egland, P. et al. “Reductive Coenzyme A-Mediated Pathway for 3-Chlorobenzoate Degradation in the Phototrophic Bacterium Rhodopseudomonas palustris”. Applied and Environmental Microbiology. 2001. Volume 67, Issue 3, p. 1396-1399.
- ↑ Egland, P. et al. “Reductive Coenzyme A-Mediated Pathway for 3-Chlorobenzoate Degradation in the Phototrophic Bacterium Rhodopseudomonas palustris”. Applied and Environmental Microbiology. 2001. Volume 67, Issue 3, p. 1396-1399.
- ↑ Egland, P. et al. “Reductive Coenzyme A-Mediated Pathway for 3-Chlorobenzoate Degradation in the Phototrophic Bacterium Rhodopseudomonas palustris”. Applied and Environmental Microbiology. 2001. Volume 67, Issue 3, p. 1396-1399.
- ↑ Harwood, C., Gibson, J. ”Anaerobic and aerobic metabolism of diverse aromatic compounds by the photosynthetic bacterium Rhodopseudomonas palustris.” Applied and Environmental Microbiology. 1988. Volume 54, Issue 3, p. 712-717.
- ↑ McGrath, J., Harfoot, C. “Reductive dehalogenation of halocarboxylic acids by the phototrophic genera Rhodospirillum and Rhodopseudomonas”. Applied and Environmental Microbiology. 1997. Volume 63, Issue 8, p. 3333-3335.
- ↑ Harwood, C., Gibson, J. ”Anaerobic and aerobic metabolism of diverse aromatic compounds by the photosynthetic bacterium Rhodopseudomonas palustris.” Applied and Environmental Microbiology. 1988. Volume 54, Issue 3, p. 712-717.
- ↑ Harwood, C., Gibson, J. ”Anaerobic and aerobic metabolism of diverse aromatic compounds by the photosynthetic bacterium Rhodopseudomonas palustris.” Applied and Environmental Microbiology. 1988. Volume 54, Issue 3, p. 712-717.
- ↑ Sharma, N. et al. “Skatole remediation potential of Rhodopseudomonas palustris WKU-KDNS3 isolated from an animal waste lagoon”. Lett Appl Microbiol. 2015. Volume 60, Issue 3, p. 298-306.
- ↑ Myung, K. et al. “Odorous swine wastewater treatment by purple non-sulfur bacteria, Rhodopseudomonas palustris, isolated from eutrophicated ponds”. Biotechnology Letters. 2004. Volume 26, Issue 10, p. 819-822.
- ↑ Xing, D. et al. “Electricity Generation by Rhodopseudomonas palustris DX-1”. Environ. Sci. Technol. 2008. Volume 42, Issue 11, p. 4146-4151.
- ↑ Yongquin, J. et al. “Isolation and Characterization of a Genetically Tractable Photoautotrophic FE(II)-Oxidizing Bacterium, Rhodopseudomonas palustris Strain TIE-1”. Applied and Environmental Microbiology. 2005. Volume 71, Issue 8, p. 4487-4496.
- ↑ Bose, A. et al. “Electron uptake by iron-oxidizing phottrophic bacteria”. Nature Communications. 2014. Volume 5, Article 3391.
- ↑ Bose, A. et al. “Electron uptake by iron-oxidizing phottrophic bacteria”. Nature Communications. 2014. Volume 5, Article 3391.
- ↑ Pant, D. et al. “A review of the substrates used in microbial fuel cells (MFCs) for sustainable energy production”. Bioresource Technology. 2010. Volume 101, Issue 6, p. 1533-1543.
- ↑ Bose, A. et al. “Electron uptake by iron-oxidizing phottrophic bacteria”. Nature Communications. 2014. Volume 5, Article 3391.
- ↑ Mehrabi,S. et al." Identification and characterization of Rhodopseudomonas spp., a purple, non-sulfur bacterium from microbial mats"."Biomolecular Engineering". 2001.Volume 18,p.49-56.
- ↑ Larimer F. et al. “Complete genome sequence of the metabolically versatile photosynthetic bacterium Rhodopseudomonas palustris”. Nature Biotechnology. 2004. Volume 22, p.55-61
- ↑ Oh, Y. et al. ”Fermentative hydrogen production by a new chemoheterotrophic bacterium Rhodopseudomonas palustris P4” International Journal of Hydrogen Energy. 2002. Volume 27, Issue 11-12, p. 1373-1379.
- ↑ Oh, Y. et al. ”Photoproduction of hydrogen from acetate by a chemoheterotrophic bacterium Rhodopseudomonas palustris P4” International Journal of Hydrogen Energy. 2004. Volume 29, Issue 11, p. 1115-1121.
- ↑ Oh, Y. et al. ”Photoproduction of hydrogen from acetate by a chemoheterotrophic bacterium Rhodopseudomonas palustris P4” International Journal of Hydrogen Energy. 2004. Volume 29, Issue 11, p. 1115-1121.
- ↑ Oh, Y. et al. ”Photoproduction of hydrogen from acetate by a chemoheterotrophic bacterium Rhodopseudomonas palustris P4” International Journal of Hydrogen Energy. 2004. Volume 29, Issue 11, p. 1115-1121.
- ↑ Pandey, A."Using reverse micelles as microreactor for hydrogen production by coupled systems of Notoc/R.palustris and Anabaena/R.palustris". "World Journal of Microbiology and Biotechnology".Volume 232. p.269-274.
- ↑ Xing, D. et al. “Electricity Generation by Rhodopseudomonas palustris DX-1”. Environ. Sci. Technol. 2008. Volume 42, Issue 11, p. 4146-4151.
- ↑ Pant, D. et al. “A review of the substrates used in microbial fuel cells (MFCs) for sustainable energy production”. Bioresource Technology. 2010. Volume 101, Issue 6, p. 1533-1543.
- ↑ Bose, A. et al. “Electron uptake by iron-oxidizing phottrophic bacteria”. Nature Communications. 2014. Volume 5, Article 3391.
Authored for BIOL 238 Microbiology, taught by Joan Slonczewski, 2016, Kenyon College.
