Community-Acquired Methicillin-Resistant Staphylococcus Aureus (CA-MRSA)
By: Tom Hardacker
Introduction
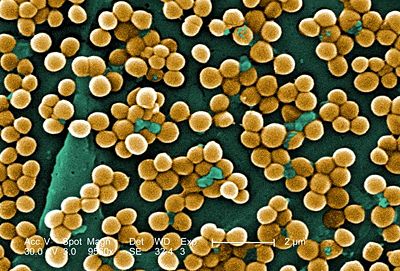
Staphylococcus aureus is a circular, anaerobic, Gram-positive bacterium that is prevalent in the nose and skin of most individuals (Figure 1). The bacteria are capable of undergoing aerobic respiration as well as anaerobic fermentation. S. aureus is a catalase-positive microbe, meaning that it produces the enzyme catalase, lending it the ability to convert hydrogen peroxide (H2O2) to water and oxygen. In many instances, the catalase test distinguishes Staphylococcus from Streptococcus. The coagulase (an enzyme that induces clot formation) test distinguishes most strains of S. aureus from other Staphylococcus species (7). The genome of S. aureus, which was mapped in 2001, contains 2.8 Mbp and approximately 2,600 open reading frames. Genes encoding resistance to antibiotics are located on plasmid vectors throughout the genome and are easily transferred to other individuals by means of horizontal gene transfer.
While the majority of individuals who are colonized by S. aureus are simply carriers, this organism can cause a wide array of illnesses if introduced into the body. Infections can range from mild skin irritation to more serious conditions such as endocarditis (inflammation of the inner heart), meningitis, pneumonia and Toxic Shock Syndrome (TSS), among others. Infections by S. aureus can also be prevalent in post-surgical wounds and due to the overuse of antibiotics; certain strains of this organism have become resistant to common treatments. For these reasons, certain strains of this organism have become increasingly problematic in hospitals and healthcare settings, as well as the general community.
Methicillin-Resistant Staphylococcus aureus (MRSA) is a strain of S. aureus that exhibits resistance to the β-lactam antibiotic methicillin (as well as other β-lactams), a common treatment for these infections. MRSA infections can be classified into two major groups: Hospital-acquired MRSA (HA-MRSA) and Community-acquired MRSA (CA-MRSA). HA-MRSA is responsible for post-operative wound infections, or infections resulting from implanted devices such as catheters, that are acquired within the healthcare setting. Typically, patients infected with HA-MRSA are immune-compromised and the resulting infections are generally more invasive. CA-MRSA typically manifests itself as skin infections, such as pimples or boils, and is classified as being acquired outside of any type of healthcare setting. These infections are typically more serious than minor skin irritation and affect otherwise healthy individuals. This article will focus on the latter form of MRSA (an in-depth article regarding HA-MRSA can be found here.
Origins of MRSA
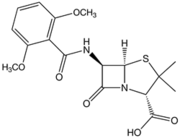
Methicillin was developed by the pharmaceutical company Beecham in 1959 in response to bacteria (namely S. aureus) that were resistant to the drug penicillin. Like penicillin, and other β-lactam antibiotics that were later developed, methicillin acts by inhibiting cell-wall synthesis in Gram-positive bacteria. This class of antibiotics binds to the transpeptidase enzyme (also called penicillin binding proteins, or PBPs), which is used by bacteria to cross-link peptidoglycan layers in the cell wall. The β-lactams competitively inhibit these enzymes and prevent these bacteria from successfully undergoing cell division. This ultimately leads to cell death and is a very effective mechanism in dealing with Gram-positive infections (8).
Methicillin was particularly effective upon its introduction into medical use because of its resistance to β-lactamases secreted by bacteria that actively degraded the β-lactam ring in penicillin. The presence of the ortho-dimethoxyphenyl group on methicillin prevents enzymatic hydrolysis of the β-lactam ring (Figure 2). However, quickly after its introduction into clinical use, S. aureus began to show resistance to methicillin. The mechanism of S. aureus resistance to methicillin is very similar to the mechanism of resistance to penicillin. Resistance hinges on the presence of the mecA gene in the chromosome. This gene encodes penicillin binding protein 2A (PBP2A), which significantly decreases methicillin’s affinity to bind to the PBP targets. The mecA gene is a part of the mobile element Staphylococcal Cassette Chromosome (SCCmec) and is easily transferred among S. aureus communities by means of plasmid transfer (3, 5).
In 1961, only two years after its introduction into clinical use, methicillin-resistant strains of S. aureus were reported in the United Kingdom. Soon thereafter, similar cases emerged in Japan, New Zealand, Australia, and the United States. Each strain carried a distinct SCCmec (Type I-V) that determine resistance capacity. Resistance to the β-lactam antibiotics is easily transferred among S. aureus individuals by means of a plasmid vector. This mechanism of transfer allows for the rapid rise of resistance among S. aureus strains. Though the cases were associated strictly with healthcare settings (HA-MRSA) and most often with immune-compromised patients, S. aureus strains developed the capacity to easily infect otherwise healthy individuals in a community setting. Certain genetic differences between HA- and CA-MRSA have been noted and are theorized to provide an ease of transfer that is associated with CA-MRSA. Though the exact mechanisms of this evolution have remained somewhat unclear, recent research has shown that CA-MRSA infections in the United States have been caused by a strain known as USA300. This same research has suggested that, though there has been minimal evolutionary divergence (based on very few single nucleotide polymorphisms), the small changes that have occurred within this strain are significant enough to cause a diversification of the USA300 strain. These findings disprove the hypothesis of convergent evolution of CA-MRSA strains in the United States (6).
Spread of Infection and Pathology
S. aureus lives harmlessly in the mucous membranes of the nose on about 33% of the general population (14). When the organism is able to penetrate the skin or mucous barriers it can lead to infection. The most common methods of spread outside of a hospital setting include: contact with pus from an infection site, skin-to-skin contact or sharing towels, clothing or athletic equipment with an infected individual. The compound hyaluronidase, which is produced by S. aureus, destroys soft tissue and allows for easier spread once inside the body.
Once an infection has occurred, the first, most common symptom is a significant abscess on the epidermis. From this abscess, proteolytic enzymes produced by S. aureus enable it to disperse throughout the body and cause secondary infections. The organism is now able to cause pneumonia and also infection of the joints, bones and heart valves. CA-MRSA strains also produce enterotoxins, which can cause serious illnesses. These toxins can be the cause of certain forms of food poisoning, Staphylococcal Scalded Skin Syndrome (SSSS), and Toxic Shock Syndrome (TSS). SSSS is a rare disease that usually affects infants by the production of an exfoliative enterotoxin. TSS is the most widely known byproduct of CA-MRSA infections. Improper use of tampons has been the cause of the vast majority of TSS cases, as the tampon can create a nutrient-rich environment for S. aureus to live. Symptoms include hypotension, fever, rash and can lead to hepatic and renal dysfunction, membrane hyperemia and thrombocytopenia (11).
Outbreaks of CA-MRSA are most common in athletic teams, prisons and military training centers and often occur in large clusters. As previously discussed, athletic teams will typically spread the infection rapidly based on skin-to-skin contact during participation in competition and/or sharing of towels or equipment in the locker room. Thus, one infected individual can rapidly pass the infection on to multiple teammates by means of this mechanism. In 2000, ten members of a Pennsylvania college football team presented with CA-MRSA infections, with seven of the members requiring hospitalization. Aforementioned risk factors explained the relatively rapid and significant spread of the infection. A fencing team reported that five of its members had been infected with MRSA. Equipment and towel sharing was reported as common. Electronic sensing wires were worn underneath clothing during competition and were not routinely cleaned. These pathways are probable culprits for the infection spread (4).
In recent years, professional teams have experienced MRSA outbreaks. Since 2003, the Cleveland Browns, Washington Redskins and St. Louis Rams have all had multiple members infected by S. aureus. There have been several notable cases of MRSA in Ohio, including several professional and high school-level incidents around the Cleveland area. In 2005, two members of the Toronto Blue Jays presented with MRSA colonies. In all cases, each sports organization sterilized locker rooms, replaced outdated whirlpools, provided surgical scrub soap to combat turf burn-induced infections and increased education regarding MRSA. A 2003 study of a MRSA outbreak in a Georgia prison revealed that the likelihood of MRSA infection was inversely associated with hand-washing and showering frequency. Individuals who were colonized by S. aureus were found to be at a higher risk for infection. Furthermore, the study found that younger ages as well as obesity are risk factors for MRSA infection. Younger age was hypothesized to result in a more active lifestyle, which in turn caused more abrasions, etc. in this setting. Obesity may lead to differing patterns of MRSA skin colonization, which can increase the potential for infection (15).
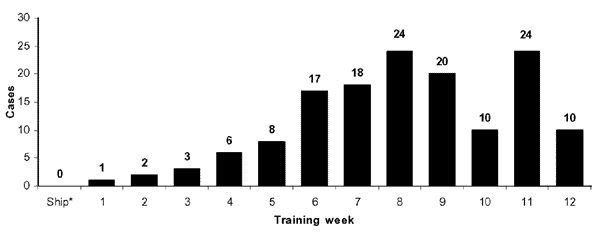
MRSA outbreaks are also problematic at military training facilities where close contact and sharing of equipment exist as significant risk factors. During a MRSA outbreak at a military training facility from October 2000 to July 2002 monthly incidence rates fluctuated from approximately 4.9 to 11 cases per 1,000 recruits. The recruits, between 17-25 years of age, most frequently presented with MRSA colonies on the extremities (73.7%). It was noted in this study that the rate of infection increased during warmer months. S. aureus infections are known to be spread more readily in warmer months (2), but other reasons for this were theorized. Some recruits complained of bug bites that lead to infection, which are seasonal. More frequently exposed skin is also a likely source of transmission in this case. In 2002, administrators placed anti-bacterial hand soaps at every recruit sink, extended time allotted for showering and cleaning, prohibited towel and razor sharing, and recommended that hand-washing be performed as frequently as possible. The outbreak subsided shortly after these measures were implemented (12).
Diagnosis and Treatment
CA-MRSA is differentiated from HA-MRSA based on an individual’s exposure to healthcare settings. If the patient has not been hospitalized (typically, the timeframe for hospitalization is up to one year), does not have any catheters or dialysis, and has not been recently admitted into a nursing home, etc. then any MRSA infection is likely community-acquired. On a microbiological level, CA-MRSA has been shown to be genetically different from HA-MRSA and the two strains can be differentiated based on lab cultures as well. These differences allow CA-MRSA to spread more easily among individuals and use skin infections as a more prevalent mechanism of infection.
Strategies for diagnosing and treating CA-MRSA with a focus on Skin and Soft Tissue Infections (SSTIs) are outlined by the Center for Disease Control (CDC). S. aureus skin infections are commonly mistaken for spider bites by patients, as they are similar in their appearance. Any condition that may potentially exist in conjunction (or as a result of) a MRSA infection, such as pneumonia, osteomyelitis, or septic arthritis, is also regarded as potentially resulting from CA-MRSA. If a physician suspects that an infection is the result of S. aureus then a culture will be taken to obtain a definite identification (a blood, urine or drainage sample may be taken depending on the type of infection). These cultures not only serve to make a definite diagnosis, but also to track individual strains of MRSA.
For furuncles, abscesses and septic joints, the primary treatment option is incision and drainage by a physician. In controlled trials, this method has been shown to be 90% effective in dealing with CA-MRSA infections. If the SSTI is progressing more rapidly, then a physician may choose to supplement the drainage of the wound with antimicrobial therapy (antibiotics, etc.). Depending upon the rate of methicillin-resistance in the MRSA colony (identified based on lab cultures) a reasonable first-line antibiotic approach would include anti-staphylococcal β-lactams and cephalosporin. Non-β-lactam antibiotics that are effective against MRSA (especially those with a high rate of methicillin-resistance within the colony) include clindamycin, the tetracyclines, trimethaprim-sulfamethoxazole (TMP-SFX), rifampin, and linezolid. The use of fluoroquinolones and macrolides is not recommended for MRSA treatment based on the rapid development of resistance to these drugs. Severe MRSA infections should be treated with intravenous antibiotics such as clindamycin, daptomycin, linezolid, quinopristin-dalfopristin, tigecycline and TMP-SFX. For the full list of diagnosis and treatment methods as outlined by the CDC, click here.
There are currently two main mechanisms that antibiotics are able to successfully inhibit S. aureus growth. One mechanism is to prevent the cross-linking of the peptidoglycan layer. Vancomycin works by this mechanism Gram-positive bacteria. As outlined by the CDC, this drug has been deemed one of “last resort” based on its adverse effects on the kidneys and hearing. Though resistance has been low in response to vancomycin (though it was originally discovered in 1958) some cases of vancomycin-intermediate S. aureus (VISA) and a handful of vancomycin-resistant S. aureus (VRSA) have been noted. The use of avoparcin in agriculture is suspected to have contributed to the rise of resistance to this antibiotic. The other mechanism is the inhibition of bacterial transcription. Linezolid and the tetracylines bind and prevent the 30S ribosomal subunit from binding the 50S subunit and initiating transcription. Clindamycin preferentially binds the 50S subunit to illicit the same result. This prevents S. aureus from synthesizing necessary proteins, which ultimately leads to cell death. Resistance to linezolid has remained extremely low (<0.5%) in the United States (9). Rifampin inhibits DNA-dependent RNA polymerase in S. aureus and also leads to cell death based on its failure to transcribe necessary proteins (10).
Mechanisms of Resistance
MRSA has gained resistance to the β-lactam antibiotics by means of enzymatic inhibition as well as altering target enzymes. Some resistant strains of S. aureus are able to synthesize an enzyme called β-lactamase, which binds to the β-lactam antibiotics and inactivate them. However, the clinical drug Augmentin is used to disable the β-lactamase enzyme and is often used in conjunction with the β-lactam antibiotics. Since β-lactams bind PBPs on the cell membrane of S. aureus, the main strategy of resistance is to alter these proteins. Once the organism alters the PBPs, the β-lactam antibiotics have a significantly decreases affinity for the substrate (PBPs). This is the main mechanism of resistance employed by the MRSA strains of S. aureus (16).
As discussed previously, the gene conferring resistance to the β-lactam antibiotics is located on the mecA gene and is easily transferred among individuals by means of a plasmid vector. This gene encodes for PBPs with altered structure and decreases the β-lactam affinity for this substrate by the mechanism described above. Vancomycin-resistance has been traced to the presence of the vanA gene, which operates by the same mechanism to prevent vancomycin and other glycopeptides antibiotics from binding their substrate on the peptidoglycan of S. aureus.
Methods of Prevention
Frequent hand-washing with soap and/or alcohol-based hand rubs has shown to be an effective way to prevent the spread of CA-MRSA. Individuals should shower immediately after any athletic event and avoid sharing bar soap, razors or towels in the process. Uniforms and athletic clothing should be washed and thoroughly dried after each game or practice. Since cuts and abrasions are the most common mechanisms of infection among athletes, they should be covered with clean bandages during each athletic activity. Any items that are shared and contact bare skin should be avoided (1).
Future Research
Recent development of antibiotics to combat MRSA infections has slowed significantly in recent decades. This trend is largely due to the exorbitant cost associated with developing, testing, and marketing new treatments. There have been new developments from pharmaceutical industries that have produced new potential antibiotic treatments. Cerexa and Telavancin are two new antibiotics that are currently undergoing testing and evaluation by the Food and Drug Administration (FDA). Both of these antibiotic treatments are effective against MRSA in lab settings; however, they use similar modes of action as prior treatments. This may potentially mean that resistance to these drugs may occur more easily.
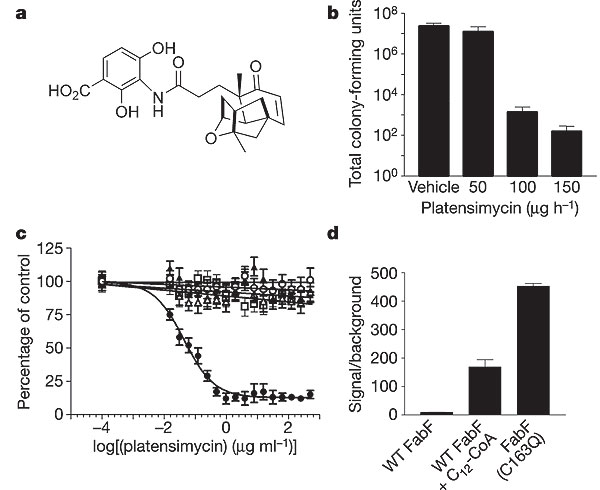
However, recent research has isolated the antibiotic platensimycin from soil samples in South America. This antibiotic is naturally produced by the organism Streptomyces platensis and has been shown to inhibit fatty acid synthesis in Gram-positive bacteria, most notably S. aureus. Platensimycin selectively binds and inhibits β-ketoacyl-(acyl-carrier-protein (ACP)) synthase I/II (FabF/B) in the biosynthesis pathway of fatty acids. Furthermore, because this antibiotic has a novel mode of action, it showed no cross-resistance to other resistant strains (MRSA, VISA, VRSA) in research settings. MRSA colonies in infected mice were eradicated with the treatment of platensimycin and this drug is currently in pre-clinical testing for use in humans. A UK-based biotechnology group, Phico Therapeutics, has presented a new study on a new treatment of MRSA infections. The SASPs are a part of a small group of proteins that are capable of inactivating bacterial DNA. The strategy employed by Phico Therapeutics is to insert the SASP gene into S. aureus by means of a delivery vector (the most promising of which is named PTSA1.2/A). Studies have shown that existing resistance had no effect on the delivery of this treatment and a greater than 3 log drop in colony counts occurred at 6 hours after treatment administration (13).
Conclusion
S. aureus infections represent a growing problem in today's society. The use of antibiotics, which lead to the subsequent antibiotic-resistance in the organism, has caused cases of infections to be increasingly difficult to treat. Rapid transmission of resistance genes on plasmid vectors has enabled resistance to common treatments to be widespread. Despite the fact that multiple classes of drugs have now become ineffective in treating MRSA, significant development of novel drugs is not occurring. New drugs that are being developed mostly rely on similar mechanisms as their predecessors, which can lead to organisms gaining resistance much quicker. Despite these facts, there are several new drugs with novel mechanisms that could significantly aid in the treatment of MRSA infections.
Though the prevalence of HA-MRSA is alarming, the rise of CA-MRSA in recent history is nearly as troublesome. Genetic elements allow CA-MRSA to rapidly spread and infect otherwise healthy individuals who have had no contact with any hospital setting. Simple cuts or abrasions are sufficient to acquire the disease, which is particularly alarming since millions of individuals suffer these risk-factors on a regular basis. Furthermore, knowledge regarding prevention is seriously lacking. This has led to poor prevention methods on sporting teams, in prisons and in military training centers. The rapid transmission of MRSA infections and the potential for them to progress into life-threatening conditions makes these situations quite dangerous. While antibiotic resistance in MRSA increases, the most effective methods of dealing with this threat remain effective prevention of infection.
References
1. Center for Disease Control Prevention of Methicillin-Resistant Staphylococcus aureus Infections. http://www.cdc.gov/ncidod/dhqp/ar_MRSA_AthletesFAQ.html#5
2. Chin J, editor. Control of Communicable Diseases Manual. 17th ed. Baltimore: United Book Press, Inc.; 2000.
3. Enright, M.; Robinson, D.; Randall, G. Feil, E.; Grundmann, H.; Spratt, B. “The evolutionary history of methicillin-resistant Staphylococcus aureus (MRSA)” Proceedings of the National Academy of Sciences. Volume 99. Pg. 7686-7692.
4. Gantz, N. “Methicillin-Resistant Staphylococcus aureus Infections Among Competitive Sports Participants --- Colorado, Indiana, Pennsylvania, and Los Angeles County, 2000—2003” Center for Disease Control. Volume 52. Pg. 793-795. http://www.cdc.gov/mmwr/preview/mmwrhtml/mm5233a4.htm
5. Jennings, Barbara. “Methicillin-Resistant Staphylococcus aureus” Smith College Molecular Biology 360.
6. Kennedy, A.; Otto, M.: Braughton, K.; Whitney, A.; Chen, L.: Mathena, B.; Miediavella, J.; Byrne, K.; Parkins, L.; Tenover, F.; Kreiswerth, B.; Musser, J.; DeLeo, F. “The evolutionary history of methicillin-resistant Staphylococcus aureus (MRSA)” The Proceedings of the National Academy of Sciences. Volume 105. Pg. 1327-1332. http://www.pnas.org/content/105/4/1327.full.pdf+html
7. Matthews, K.R.; Roberson, J.; Gillespie, B.E.; Luther, D.A.; Oliver, S.P. “Identification and Differentiation of Coagulase-Negative Staphylococcus aureus by Polymerase Chain Reaction” The Journal of Food Protection, Volume 60. June 1997. Pg. 686-688(3) http://www.ingentaconnect.com/content/iafp/jfp/1997/00000060/00000006/art00015
8. Mitscher LA. Antibiotics and Antimicrobial Agents. Foye's Principles of Medicinal Chemistry. Philadelphia: Lippincott Williams & Wilkins; 2002
9. Saager B, Rohde H, Timmerbeil BS, et al (September 2008). "Molecular characterisation of linezolid resistance in two vancomycin-resistant (VanB) Enterococcus faecium isolates using Pyrosequencing". Eur J Clin Microbiol Infect Dis 27 (9): 873–8. doi:10.1007/s10096-008-0514-6. PMID 18421487.
10. Sigma Aldrich. “Antibiotic Mechanisms of Action.” http://www.sigmaaldrich.com/life-science/biochemicals/antibiotic-explorer/Mechanism-of-Action.html
11. Tolan, R. “Staphylococcus Aureus Infection” Infectious Disease. http://emedicine.medscape.com/article/971358-overview
12. Turabelidze, G.; Lin, M.; Wolkoff, D.; Dodson, D.; Gladbach, S.; Zhu, B. “Personal Hygiene and Methicillin-Resistant Staphylococcus Aureus Infection.” Center for Disease Control. Volume 3. http://www.cdc.gov/ncidod/eid/vol10no5/03-0604.htm
13. Wang, et. al. “Plastensimycin is a Selective FabF Inhibitor with Potent Antibiotic Properties” Nature. Volume 441. Pg. 358-361. http://www.nature.com/nature/journal/v441/n7091/abs/nature04784.html
14. Whitt, Dixie D.; Salyers, Abigail A.. "14". Bacterial Pathogenesis: A Molecular Approach (2nd edition ed.). USA: ASM Press. ISBN 1-55581-171-X.
15. Zindermann, C.; Conner, B.; Malakooti, M.; LaMar, J. ; Armstrong, A. ; Bonker, B. “Community-Acquired Staphylococcus aureus Infections Among Military Recruits” Center for Disease Control. Volume 5.
16. Zoidis, J. “Mechanisms of Resistance, Emergent Strains, and Methods of Control. RT For Decision Makers in Respiratory Care. December 1998.
Edited by student of Joan Slonczewski for BIOL 238 Microbiology, 2009, Kenyon College.
