Desulfovibrio vulgaris: Difference between revisions
| Line 28: | Line 28: | ||
==Genome structure== | ==Genome structure== | ||
Desulfovibrio vulgaris has a genome that is 3,570,858 base pairs in length. The chromosome has a 64.2% guanine and cytosine concentration. Remarkably, the chromosome has 1,894 similar to known protein coding genes in a predicted 3,395 total coding sequences. These genes code for an intricate network of c-type cytochromes that connect multiple periplasmic hydrogenases and formate dehydrogenases (4). Additionally, the sequencing showed the presence of a phosphotransferase system (PTS) similar to those found in other Gram (-) and Gram (+) bacteria that suggest D. vulgaris may have the potential to use mannose type sugars. However evidence that this bacterium uses sugars has not yet been confirmed even though the presence of most of the necessary proteins has been confirmed. In the strain D. vulgaris Hildenborough many proteins were found that are used in glycolysis. The table below does the protein and the gene it is coded from. | |||
Glucose phosphate isomerase (pgi) | |||
Phosphofructokinase (pfkA) | |||
Fructose bisphosphate aldolase (fba) | |||
Triose phosphate isomerase (tpiA) | |||
Glyceraldehydes-3-phosphate dehydrogenase (gap-1, gap-2) | |||
3-Phosphoglycerate kinase (pgk) | |||
Phosphoglycerate mutase (gpm ,gpmA) | |||
Enolase (eno) | |||
Pyruvate kinase (pyk) | |||
The genome sequencing illustrates the possibility of hydrogenases and cytochromes that remove the hydrogens from metals (3). This is substantiated by the presence of genes coding for 27 methyl-accepting chemotaxis proteins that include proteins DcrA andDcrH that sense gradients of oxic/anoxic interfaces (3). | |||
Another component of the genome that is being researched are the genes that code for proteins that reduce oxygen are found on the same chromosome. These genes are superoxide reductase (Sor), rubredoxin (Rub), and rubredoxin:oxygen oxidoreductase (Roo) (9). | |||
==Cell structure and metabolism== | ==Cell structure and metabolism== | ||
Revision as of 20:22, 29 August 2007
A Microbial Biorealm page on the genus Desulfovibrio vulgaris
Classification
Higher order taxa
Cellular organism: Bacteria (Domain): Proteobacteria(Phylum): delta/epsilon subdivisions (Sub-phylum): Deltaproteobacteria (Class): Desulfovibrionales (Order): Desulfovibrionaceae (Family) [Others may be used. Use NCBI link to find]
Species
|
NCBI: Taxonomy |
Desulfovibrio (Genus): Desulfovibrio vulgaris (Species)
Description and significance
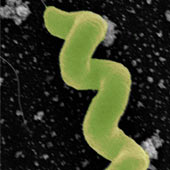
Desulfovibrio vulgaris is a Gram-negative , anaerobic, non-spore forming, curved rod-shaped bacteria, that can be found in soil, animal intestines and feces, and fresh and salt water. (1) It is able to thrive in many of these environments as a result of cellular modifications as well as metabolic enhancements that has allowed this species to live in terrestrial and aquatic habitats. Sulfur-reducing bacteria (SRB), such as D. vulgaris are generally found in anoxic sub-surfaces like areas formed from microbial decomposition as well as sediments and bottom waters. (5). D. vulgaris Hildenborough was discovered in clay soil near Hildenborough, Kent, United Kingdom in forests and grazing fields. Many of the bacteria found in the rhizosphere where roots from plants releasing organic material (6).
The strain many researchers use come from Hildenborough, UK that was first isolated in 1946 from clay. The carbon sources it can use are acetate, pyruvate, formate, and certain primary alcohols (2,3). Desulfovibrio oxidize their energy sources to acetate, excreting this as their end product.
Desulfovibrio species are characterized by contain desulfoviridin. This genus can grow easily on a sulfate-lactate medium in the absence of oxygen as a result of metabolic evolutionary traits that have been since studied and isolated by biotechnicians.
Desulfovibrio vulgaris Hildenborough is a model organism for studying the energy metabolism of sulfate-reducing bacteria and for understanding the related economic impacts, including bio-corrosion of metal infrastructure and bioremediation of toxic metal ions. It is also known as the “petroleum pest” since it is known to cause a lot of damage to equipment used for drilling and storing oil. What makes this sulfate-reducing bacteria unique is that it can cause the enzymatic reduction of Cr(VI) into a nontoxic form. Furthermore it makes use of hydrogenase activity and cytochrome c3 as the reductase. What separates this reducer is that it can function in high concentration of toxicity as well as in the presence of nickelous chloride, zinc chloride, magnesium sulfate, vanadyl sulfate, and many more (4).
Genome structure
Desulfovibrio vulgaris has a genome that is 3,570,858 base pairs in length. The chromosome has a 64.2% guanine and cytosine concentration. Remarkably, the chromosome has 1,894 similar to known protein coding genes in a predicted 3,395 total coding sequences. These genes code for an intricate network of c-type cytochromes that connect multiple periplasmic hydrogenases and formate dehydrogenases (4). Additionally, the sequencing showed the presence of a phosphotransferase system (PTS) similar to those found in other Gram (-) and Gram (+) bacteria that suggest D. vulgaris may have the potential to use mannose type sugars. However evidence that this bacterium uses sugars has not yet been confirmed even though the presence of most of the necessary proteins has been confirmed. In the strain D. vulgaris Hildenborough many proteins were found that are used in glycolysis. The table below does the protein and the gene it is coded from.
Glucose phosphate isomerase (pgi)
Phosphofructokinase (pfkA)
Fructose bisphosphate aldolase (fba)
Triose phosphate isomerase (tpiA)
Glyceraldehydes-3-phosphate dehydrogenase (gap-1, gap-2)
3-Phosphoglycerate kinase (pgk)
Phosphoglycerate mutase (gpm ,gpmA)
Enolase (eno)
Pyruvate kinase (pyk)
The genome sequencing illustrates the possibility of hydrogenases and cytochromes that remove the hydrogens from metals (3). This is substantiated by the presence of genes coding for 27 methyl-accepting chemotaxis proteins that include proteins DcrA andDcrH that sense gradients of oxic/anoxic interfaces (3).
Another component of the genome that is being researched are the genes that code for proteins that reduce oxygen are found on the same chromosome. These genes are superoxide reductase (Sor), rubredoxin (Rub), and rubredoxin:oxygen oxidoreductase (Roo) (9).
Cell structure and metabolism
Describe any interesting features and/or cell structures; how it gains energy; what important molecules it produces.
Ecology
Describe any interactions with other organisms (included eukaryotes), contributions to the environment, effect on environment, etc.
Pathology
How does this organism cause disease? Human, animal, plant hosts? Virulence factors, as well as patient symptoms.
Application to Biotechnology
Does this organism produce any useful compounds or enzymes? What are they and how are they used?
Current Research
Enter summaries of the most recent research here--at least three required
References
Edited by student of Rachel Larsen