Drug Resistance of P. aeruginosa: Difference between revisions
| Line 27: | Line 27: | ||
==References== | ==References== | ||
1. | 1. [http://en.wikipedia.org/wiki/Pseudomonas_aeruginosa#cite_note-18Pseudomonas Aeruginosa." Wikipedia. Wikimedia Foundation, 12 Apr. 2013. Web. 01 Dec. 2013.] | ||
2. Fajardo, Alicia, and Jose L. Martinez. "Chapter 8: Antibiotic Resistance in Pseudomonas." Pseudomonas: Genomics and Molecular Biology. By Pierre Cornelis. Norfolk, UK: Caister Academic, 2008. N. pag. Print. | 2. Fajardo, Alicia, and Jose L. Martinez. "Chapter 8: Antibiotic Resistance in Pseudomonas." Pseudomonas: Genomics and Molecular Biology. By Pierre Cornelis. Norfolk, UK: Caister Academic, 2008. N. pag. Print. | ||
Revision as of 04:28, 7 December 2013
Introduction
Pseudomonas aeruginosa is a type of bacterium that has the ability to develop resistance to antibiotics rather rapidly over several generations. This resistance present in some strains makes P. aeruginosa very difficult to treat once a host, such as a human or other animal, is infected. Adding to its ability to develop drug resistance is its resilience that allows it to thrive in various environments, especially medical environments and moist environments containing water. It is commonly a hospital-acquired infection; though there is a distinction between its colonization and its infection.1 It is gram-negative, rod shaped, and aerobic. Additionally it has the ability to catalyze numerous organic molecules and mutate to adapt to its environmental nutritional conditions.
Other examples:
Bold
Italic
Subscript: H2O
Superscript: Fe3+
Infection
Pseudomonas aeruginosa is an opportunistic pathogen, meaning that in order to infect its host it takes advantage of any weakness in the host’s defenses. It can infect plants or animals, including humans. Although it is a normal bacterium that exists on human skin without causing harm, it becomes damaging when the immune system is compromised or an open wound or burn facilitates its entrance to the body. The most serious infections of P. aeruginosa occur in deep tissues such as blood or bone, in patients with compromised immune systems or cystic fibrosis. It is naturally resistant to common antibiotics due to chromosomally encoded genes.1 Furthermore, it has specific characteristics that contribute to its virulence, including flagella to promote motility, pili to attach to tissue surfaces, and lipopolysaccharide to act as an endotoxin.
It can grow on minimal media, requires little extra source of nutrition, thus enhancing its survival in environments that would otherwise not promote extensive bacterial growth. In clinical environments especially it can proliferate on surfaces and has become a major cause of nosocomial infections. When P. aeruginosa replicates enough, it forms a biofilm that attaches to a surface and is difficult to destroy. Infection occurs when P. aeruginosa adheres to a tissue surface and then reproduces to the point of having a mass capable of infection, and finally damages the tissue through the toxins it produces. Virulent factors, such as exotoxin A, work to colonize the host cell by inhibiting protein synthesis and attacking the structural proteins of the cell.
Treatment
P. aeruginosa is resistant to disinfectants as well as antibiotics. Resistance develops as the bacterium is exposed to different kinds of medication in a similar fashion as a MRSA infection: its low susceptibility combined with rapid reproduction result in continual adaptation to the antibodies and high bacterial diversity. It acquires resistance genes and specific mutations that enhance its survival in the presence of antibiotics. Its genome was sequenced completely in 2000 and was recorded as having 6.3 million base pairs. Such a notably large genome encodes many proteins contributing to its versatility in adaptation, virulence function, and antimicrobial resistance. Both size and complexity of genome lead to P. aeruginosa’s ability to survive and resist antibiotics, and its outer membrane has low permeability to antibiotics as well.
When an infection in human tissue is acquired in a clinical setting and treated with an antibiotic, quorum sensing among the bacterial population allows the cells to communicate with each other population molecular weight information. This gives them an indication of where the biofilm’s outermost layer is being destroyed by antibiotics so that inner layers increase their growth, and the infection persists.
Specific antibodies can target the characteristics that assist in P. aeruginosa’s ability to infect, and due to the multifactorial nature of the bacteria’ virulence, there are multiple pathways available to decrease the spread and intensity of infection. Anti-LPS (lipopolysaccharide) antibodies can prevent the commencement of infection and have been shown to decrease tissue damage. Antiflagellar antibodies can disable the bacterium’s ability to move, thus severely disrupting its ability to invade the host and halting infection. Although these antibodies have potential to develop into a vaccine for human, it “cannot be stated with certainty” which singular or collection of antibodies best protect against infection.
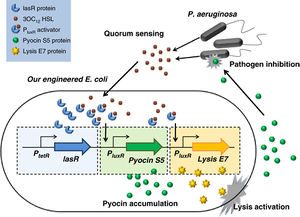
Vaccine development is being studied as a method of treatment, specifically whole-cell vaccines that utilize heat-killed strains of P. aeruginosa. Also the combating of bacterium with bacterium—in this case, the potential manipulation of E. coli bacteria to destroy P. aeruginosa bacterium—has shown promise is recent studies.
In 2011, scientists of Nanyang Technological University located in Shanghai, China, have begun the initial stages of utilizing engineered E. coli to target and destroy P. aeruginosa with its own toxin. This is achieved by constructing an E. coli with three specific genes in its genome: one to produce the same pyocin as P. aeruginosa, one to detect the presence of P. aeruginosa through the signals used in quorum sensing, and a third to induce cell lysis to release the toxins from E. coli. Once the pyocin is released from the lysed E. coli it kills the P. aeruginosa cells as well. The issue remains in the potential for E. coli to become a pathogen as well and being to infect the host along with P. aeruginosa.
Conclusion
Pseudomonas aeruginosa presents a great challenge in the clinical environment because of its antibiotic resistance and prevalence of infection in patients with open wounds and compromised immune systems. Its biofilms are difficult to destroy and its survival persists within and without its host. Multiple methods of treatment are being explored with varying effectiveness and potential for development against human infection, including vaccinations using the bacterial cells themselves and antibodies targeting virulence-promoting factors of the bacteria’s structure. Most often, however, P. aeruginosa is a long-term infection in already vulnerable, hospitalized patients, the worst possible environment for such an opportunistic pathogen to dominate. Similar antibiotic-resistant infections, such as the more commonly known MRSA infection, are termed “superbugs” due to their strength and persistence from their ability to evolve greater resistance to the majority of antibiotics used for treatment.
References
1. Aeruginosa." Wikipedia. Wikimedia Foundation, 12 Apr. 2013. Web. 01 Dec. 2013.
2. Fajardo, Alicia, and Jose L. Martinez. "Chapter 8: Antibiotic Resistance in Pseudomonas." Pseudomonas: Genomics and Molecular Biology. By Pierre Cornelis. Norfolk, UK: Caister Academic, 2008. N. pag. Print.
3. Stover, C. K., and X. Q. Pham. "Complete Genome Sequence of Pseudomonas Aeruginosa PAO1, an Opportunistic Pathogen." Nature.com. Nature Publishing Group, 10 July 2000. Web. 04 Dec. 2013.
4. Todar, Kenneth, PHD. "Pseudomonas." Pseudomonas. N.p., n.d. Web. 05 Dec. 2013.
5. Mesaros, N., and P. Nordmann. "Pseudomonas Aeruginosa: Resistance and Therapeutic Options at the Turn of the New Millennium." Wiley Online Library. N.p., 31 Jan. 2007. Web. 04 Dec. 2013.
6. Chang, Matthew W., and Nazinin Saeidi. "Engineering Microbes to Sense and Eradicate Pseudomonas Aeruginosa, a Human Pathogen." Nature.com. Nature Publishing Group, 16 Aug. 2011. Web. 04 Dec. 2013.
Edited by [Elizabeth Eder], student of Joan Slonczewski for BIOL 116 Information in Living Systems, 2013, Kenyon College.