Early gut colonization and type 1 diabetes mellitus
Caesarean section, commonly known as C-section, is a surgical procedure that allows pregnant women to deliver one or more babies through incisions made in the abdomen and uterus. In the case that vaginal delivery poses a serious threat to the health of either the mother or the child, C-section is a successful and often necessary alternative to vaginal delivery. From 1996 to 2009, the rate of C-section among all gestational age groups increased by more than 40 percent in the United States alone, a trend that has benefited from an increase in social tolerance of surgical intervention, monetary incentives on the part of healthcare providers and limited patient awareness of short and long-term risks associated with the procedure.1,2
Recent medical interest in the human gut flora, or gut microbiota, has yielded multiple studies that suggest that mode of delivery heavily impacts early colonization of the infant gut microbial community and alters the long-term composition and diversity of the gut microbiota. Furthermore, a growing body of evidence links both aberrance of the gut microbiota and mode of delivery with increased risk for immune disorders including allergy, gastrointestinal disease, and type 1 diabetes mellitus.3 Coincidently, the American Diabetes Association reported a 23 percent increase in the number of reported cases of type 1 diabetes from 2001 to 2009, a trend that may be driven by a variety of environmental factors.4 As a potential risk factor for type 1 diabetes, C-section and consequent aberrance of the gut microbiota have recently received scrutiny in the context of early and late immune health.
Modes of Delivery
Vaginal Delivery
When an infant passes through its mother’s birth canal during vaginal delivery, it is exposed to its first non-sterile environment outside the uterus via direct contact between the vagina and the infant's own mucous membranes. The vagina is typically colonized by a wide variety of anaerobic bacteria and is dominated by several species of lactic acid bacteria.5,6 During delivery, infants also come into contact with the microbes that colonize their mothers’ feces as a result of the proximity of the birth canal and the anus.5 Direct contact with the mother’s lower genital tract and anus initiates colonization of the infant’s skin and oral mucosa, and eventually of the infant’s intestinal tract, which is first dominated by facultative anaerobes including enterobacteria and lactobacilli, and then dominated by other anaerobes including Bifidobacteria, Bacteroides, Clostridia and Eubacteria.7 Following expulsion from the birth canal, infants are continuously exposed to human oral and cutaneous microbes through mother-infant contact and to other microbes in the surrounding environment, and they continue to ingest gut-colonizing microbes during the breastfeeding process and after transitioning to a solid diet. By two years of age, most healthy infants show a gut microbial composition similar to that of a typical healthy adult.5
Caesarean Section
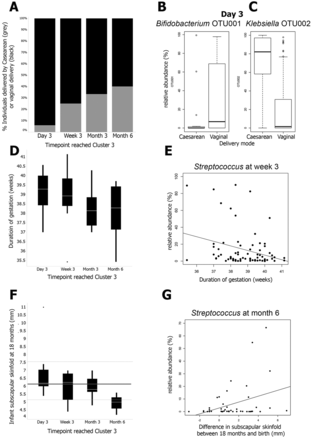
During delivery by C-section, the infant bypasses the birth canal and is directly removed from the mother’s body through incisions made in her abdomen and uterus.8 As opposed to initial microbial exposure via direct vaginal and anal contact, infants delivered by C-section are initially exposed to microbial isolates from the medical and nursing staff, surgical equipment and other elements of the immediate external environment.5 As a result, early microbial composition—including diversity and total cell count—of the skin and oral/nasal mucosa in infants delivered by C-section differs greatly from the early microbiota of infants delivered vaginally, as shown in Figure 2 on the right. Directly following delivery, the skin and oral/nasal mucosal microbiota of vaginally delivered infants resemble the mother’s vaginal microbiota and are dominated by microbes of the Lactobacillus genus. The microbiota of infants delivered by C-section closely resembles the microbiota of skin surfaces and is dominated by Staphylococcus directly following delivery.9
Gut Microbiota Aberrance
Initial colonization of the skin and mucous membranes is closely linked with the successive colonization of the infant intestinal tract, whose composition is highly affected by the initial composition of the mucosal microbiota. During the first month of life, the infant gut microbiota is composed predominantly of Bifidobacteria, a genus of Gram-positive anaerobes that are found in moderate numbers in the lower female genital tract. In a 2007 study that controlled for variant breastfeeding among test subjects, a group of researchers from Finland found that the number of Bifidobacteria in vaginally delivered infants was 1,300-fold higher than in infants delivered by C-section at one month of age.7 The same study reported significantly hampered mucosal immunity in C-section infants at 6 months of age, which suggests that early composition of the gut microbiota may be related to immune health.
Immunomudulatory effects of Bifidobacteria
As a demonstrated key component of the commensal gut microbiota, the genus Bifidobacterium is of great interest in the context of the immune system. The symbiotic interaction between Bifidobacteria and the mammalian immune system can be explained in part by the “old friends" hypothesis, which proposes that mammalian coevolution with microorganisms in the host gut allows the host immune system to recognize the once-foreign bacteria as “self.” Meanwhile, these microbes interact with the host digestive and immune systems, aiding in proper digestion of a wide variety of carbohydrates and priming immune system members for immunoregulation.21 Although specific metabolic characterization of intestinal Bifidobacteria within the larger immune system is still an emerging science, recent studies have demonstrated the significant anti-inflammatory effects of the Bifidobacteria and its critical role in both the homeostasis of the intestine and in dendritic cell modulation.15
Characterization of Bifidobacteria
Phylum: Actinobacteria
Class: Actinobacteria
Order: Bifidobacteriales
Family: Bifidobacteriaceae
Genus: Bifidobacterium16
The genus Bifidobacterium is a group of Gram-positive, rod-shaped anaerobic microbes that typically inhabit the gastrointestinal tract (GIT) and urogenital tract of mammals. These bacteria thrive in temperatures between 36 and 38 oC and in a pH range of 6.5 to 7.17 Bifidobacteria that reside in the human body are typically isolated from the feces of adults and infants.16 Accordingly, Bifidobacteria thrive in the mammalian GIT, which is supplied with a constant source of carbohydrates, has a pH that remains within the range of 5.7 to 7.4, and has a temperature range of 37 and 42 oC.18 Bifidobacteria express a unique fructose-6-phosphate phosphoketolase pathway that drives the fermentation of carbohydrates—most notably the non-digestible oligosaccharides commonly found in breast milk. This fermentation pathway, termed the “bifid shunt,” allows these bacteria to ferment a wide variety of carbon sources and results in the formation of acetic and lactic acids.16 In most healthy adults, Bifidobacteria predominate in the gut microbiota (108 - 109 cells/g of intestinal content) for the host’s entire lifetime.
As discussed previously, normal colonization of the mammalian gut with Bifidobacteria is reliant on vaginal delivery. Independent of this correlation, a 2009 study that compared the bacterial genera in the guts of diabetes resistant (BB-DR) and diabetes-prone (BB-DP) biobreeding rats found a higher abundance of the probiotic Lactobacillus and Bifidobacterium genera in BB-DR rats, suggesting that a lack of these bacteria in the rodent gut microbiota is associated with the onset of type 1 diabetes.12 A 2013 study comparing the gut microbiota of children with type 1 diabetes and the microbiota of healthy children--all delivered vaginally and breastfed--also found a significant decrease in the number of Lactobacilli and Bifidobacteria in children with diabetes.25 The symbiotic relationship between Bifidobacteria and its host may thus be a key factor in the prevention of autoimmune diabetes, and the following sections propose a possible mechanism of interaction between these bacteria and the host immune system.
Pathophysiology of type 1 diabetes
Type 1 diabetes mellitus (T1DM), commonly known as type 1 diabetes, is a metabolic autoimmune disease that develops in early childhood or adolescence. The disease is characterized by chronic hyperglycemia, which results from defects in insulin production due to autoimmune destruction of insulin-producing beta cells in the pancreas.10 The precise cause of T1DM is so far unknown, although individuals with a family history of the disease are known to be at higher risk for developing T1DM. Beyond genetic susceptibility, T1DM is currently thought to be triggered by one or more environmental factors.11
Within the pancreas, regions of endocrine cells called the islets of Langerhans are responsible for producing hormones like glucagon, insulin and amylin. Beta cells, which are one of the five secretory cells located within the islets of Langerhans, are responsible for storing and secreting insulin and amylin directly into the bloodstream. When beta cells become damaged or undergo turnover, they release autoantigens which, in healthy individuals, are not acted upon by the immune system.14 However, when these autoantigens are received by antigen-presenting cells within the pancreas that recognize the autoantigens as foreign bodies, the antigen-presenting cell engulfs the autoantigen and presents the antigen particles on its surface. Antigen presentation activates immune T cell differentiation into T helper cells, which are then able to differentiate into Th1 and Th2 cells that produce pro-inflammatory and anti-inflammatory cytokines, respectively. Th1 cytokines activate cytotoxic T cells, which destroy beta cells within the islets and suppress insulin production and excretion by the host.13 When Th1 differentiation outweighs Th2 differentiation, as is the case in type 1 diabetes, beta cell destruction is accelerated.19
The phenotype of the antigen-presenting cells that initiate the differentiation cascade of T cells governs the nature of the immune response. In particular, different phenotypes of dendritic cells have been shown to produce varying inflammatory and tolergenic responses.28 As some of the first antigen-presenting cells to infiltrate the islets during an autoimmune response, dendritic cells that produce excessive inflammatory (Th1) responses are likely culprits of beta cell autoimmunity.
Dendritic Cell Modulation
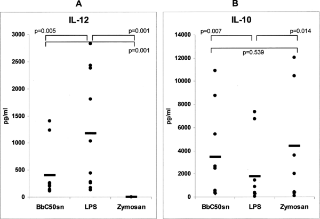
The specific antigen recognition and tolerance profile of each active dendritic cell is highly dependent on a process called maturation, which is influenced both by the stimuli provided by the engulfed autoantigen and by other members of the immune environment that interact with antigen-presenting cells. A 2006 in vitro study of the effects of Bifidobacterium breve on DC maturation showed that the supernatent of a Bifidobacterium breve C50 strain (BbC50SN) upregulated costimulatory molecule expression by activated DCs (BbC50SN-DCs), increased the production of the anti-inflammatory cytokine IL-10 (as shown in Figure 3 on the left), and increased the average lifespan of DCs.23 The same study found that BbC50SN downregulated the expression of pro-inflammatory cytokine IL-12, which is involved in the Th1 pro-inflammatory pathway linked to certain autoimmune disorders.
TLR2 Mechanism
Toll-like receptors are transmembrane proteins in antigen-presenting cells that recognize a variety of microbe-associated molecular patterns (MAMPs) that differentiate bacterial cells from host cells. TLRs are key members of the innate immune system that are responsible for mounting an immune response against invading pathogens. However, TLRs also able to recognize the MAMPs of non-pathogenic bacteria within the body, and recent evidence suggests that symbiotic gut microbes like Bifidobacteria interact with TLRs to modulate dendritic cell maturation and ultimately to modulate the host immune response.
Among the 11 currently identified TLR pathways, the TLR2 pathway has been shown to play a key role in the gut-microbiota cross-talk. Present in the cell membranes of dendritic cells, the protein TLR2 recognizes MAMPs of Gram-positive bacteria, including BLP (Bacterial Lipoprotein), PGN (Peptidoglycan) and lipotechoic acids, and binds to these bacterial components, triggering an immune response against the bacterial invader.19 As Gram-positive bacteria, Bifidobacteria contain a thick peptidoglycan layer in the cell wall, whose fragments are recognized by the surface TLRs of dendritic cells and act as TLR2 ligands.
When Bifidobacteria MAMPs act as ligands for TLR2 receptors, they induce a constant background of low-grade regulatory T cell (Treg) upregulation. Upon binding to TLR receptors, symbiotic gut bacteria like Lactobacilli present their DNA to APCs in a package that identifies them as “self.”22In turn, these Treg cells downregulate pro-inflammatory processes, maintaining a healthy population of “old friends” within the gut and increasing tolerance to “self.” The exact molecular mechanism behind this package presentation and the subsequent modulation of dendritic cell phenotype is still under debate, but there is significant evidence that the probiotic microorganisms like Bifidobacteria orchestrate dendritic cell maturation and activation upon binding with TLR2 on dendritic cells.20
Probiotic prevention and treatment of type 1 diabetes
Despite the observed links between caesarean section, gut microbiota aberrance and subsequent risk of developing autoimmune diabetes, many mothers have no choice but to deliver by C-section due to chronic maternal health issues, fetal distress during labor, or any number of complications that make vaginal birth unsafe for either mother or child. Understanding the bacteria-host interactions, or lack thereof, that comprise the core of autoimmunity is therefore a crucial first step towards developing targeted probiotic therapies for the prevention and treatment of immune disorders related to gut microbiota aberrance.
Rodent Model
In 2005, one the first controlled type 1 diabetes-specific studies of probiotic treatment of immune disorders was published in the journal Diabetologia. This study tracked the blood glucose levels, cytokine expression and beta cell destruction rate in non-obese diabetic (NOD) mice who were orally administered the probiotic formulation VSL#3 starting from 4 weeks of age. VSL#3 consists of 3×1011/g viable lyophilised bacteria, including Bifidobacteria (B. longum, B. infantis and B. breve), 4 species of Lactobacilli and 2 species of Streptococci. In comparison to the levels observed in the control group, VSL#3 administration significantly increased in the production of cytokine IL-10 and decreased beta cell destruction in NOD mice, delaying the onset of and decreasing the incidence of type 1 diabetes in probiotic-treated individuals.24
The TEDDY Study
In 2004, a study named the The Environmental Determinants of Diabetes in the Young (TEDDY) was launched in Finland, Sweden, the United States (Colorado, Georgia, and Washington State), and Germany. This ongoing project, led by Dr. Ulla Uusitalo, a nutritionist at the University of South Florida, Tampa, is testing the hypothesis that early exposure to probiotics reduces a child's risk of developing beta cell autoimmunity.
To test this hypothesis, researchers collected blood specimens from a sample group of 8676 infants with type 1 diabetes-associated alleles and tested for the presence of one or more islet autoantibodies in the blood plasma. They regularly collected information about the mother's lifestyle during pregnancy and various factors impacting the child, including illness, medications, breastfeeding, and probiotic supplementation (which primarily consisted of Lactobacilli and Bifidobacteria). 7468 of these infants were exposed to probiotics before 1 month of age, between 1 and 3 months, or between 3 and 12 months of age. The researchers adjusted the findings by country and for several influencing factors, including mode of delivery, breastfeeding and autoimmune susceptibility genotype.
When compared with the infants who were first exposed to probiotics after 12 months of age or not at all, the group that was exposed to probiotics before 1 month of age showed the highest reduction in beta cell autoimmunity (p=0.22). The group that was exposed to probiotics between 1 and 3 months of age also showed a reduction in autoimmunity (p=0.097). Together, the two groups showed a 33% reduction in beta cell autoimmunity (p=0.005). Not only do these findings indicate that probiotics containing Lactobacilli and Bifidobacteria decrease the incidence of type 1 diabetes in children, but they also suggest that early probiotic administration is the most effective mode of treatment. The commensal gut microbiota is therefore a crucial aspect of healthy development that regulates and ultimately governs short-term and long-term immune and autoimmune responses.26
Further Reading
Although this page primarily discusses a toll-like receptor mechanism of action of the Bifidobacteria-host cross talk within the gut, a recent article published in the journal of Applied and Environmental Microbiology proposes a TLR-independent mechanism of dendritic cell modulation by Bifidobacterium bifidum that is mediated by the lytic peptidoglycan enzyme TgaA.
Furthermore, a study published in Physiological Reports in March 2015 examines the interactions between Bifidobacteria and the cells of the intestinal epithelium in the context of inflammatory bowel disease, an immune system disorder that has been linked to gut microbiota aberrance. According to this study, Bifidobacteria may exert their dominant immunoregulatory effects by restoring and regulating tight junction barrier function. Because a loss of intestinal barrier function is shown to increase one's risk for developing type 1 diabetes, the findings of this article provide a promising addition and/or alternative to the TLR2 mechanism of immunoregulation by Bifidobacteria.27
References
1. Osterman MJK, Martin JA. "Changes in cesarean delivery rates by gestational age: United States, 1996–2011." NCHS data brief, no 124. Hyattsville, MD: National Center for Health Statistics. 2013. Web. http://www.cdc.gov/nchs/data/databriefs/db124.htm#x2013;2011</a
2. "Why Is the National U.S. Cesarean Section Rate So High?" Childbirth Connection. N.p., n.d. Web. http://www.childbirthconnection.org/article.asp?ck=10456
3. Cho, C. E., & Norman, M. “Cesarean section and development of the immune system in the offspring.” American Journal of Obstetrics and Gynecology 208.4 (2013): 249–54. Web. http://www.ncbi.nlm.nih.gov/pubmed/22939691
4. "Growing Number of Autoimmune Disease Cases Reported." Newswise. American Autoimmune Related Diseases Association, 21 June 2012. Web. http://www.newswise.com/articles/growing-number-of-autoimmune-disease-cases-reported
5. Mackie RI, Sghir A, Gaskins HR. "Developmental microbial ecology of the neonatal gastrointestinal tract." Am J Clin Nutr 69 (1999): 1035S–1045S. Web. http://ajcn.nutrition.org/content/69/5/1035s.short
6. Vásquez, Alejandra et al. “Vaginal Lactobacillus Flora of Healthy Swedish Women.” Journal of Clinical Microbiology 40.8 (2002): 2746–2749. PMC. Web. http://www.ncbi.nlm.nih.gov/pmc/articles/PMC120688/
7. Huurre, Anu, et al. "Mode of delivery-effects on gut microbiota and humoral immunity." Neonatology 93.4 (2008): 236. Web. http://www.ncbi.nlm.nih.gov/pubmed/18025796
8. "C-Section: What Can You Expect for Recovery?" MedicineNet. N.p., n.d. Web. http://www.medicinenet.com/c-section_cesarean_birth/article.htm
9. Dominguez-Bello MG, Costello EK, Contreras M, Magris M, Hidalgo G, Fierer N & Knight R. “Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns.” PNAS 107 (2010): 11971–11975. Web. http://www.pnas.org/content/107/26/11971.full
10. "About diabetes". World Health Organization. Web. http://www.who.int/diabetes/action_online/basics/en/
11. Belle, T. L. Van, K. T. Coppieters, and M. G. Von Herrath. "Type 1 Diabetes: Etiology, Immunology, and Therapeutic Strategies." Physiological Reviews 91.1 (2011): 79-118. Web. http://physrev.physiology.org/content/91/1/79.long
12. Roesch, Luiz Fw, Graciela L. Lorca, George Casella, Adriana Giongo, Andres Naranjo, Arianna M. Pionzio, Nan Li, Volker Mai, Clive H. Wasserfall, Desmond Schatz, Mark A. Atkinson, Josef Neu, and Eric W. Triplett. "Culture-independent Identification of Gut Bacteria Correlated with the Onset of Diabetes in a Rat Model." The ISME Journal 3.5 (2009): 536-48. Web. http://www.ncbi.nlm.nih.gov/pubmed/19225551
13. Yoon J.-W., Jun H.-S." Autoimmune destruction of pancreatic β cells." American Journal of Therapeutics 12.6 (2005):580–591. http://www.ncbi.nlm.nih.gov/pubmed/16280652
14. Poletaev AB, Churilov LP, Stroev YI, Agapov MM. (2012). "Immunophysiology versus immunopathology: Natural autoimmunity in human health and disease." Pathophysiology (2012). Web. http://www.pathophysiologyjournal.com/article/S0928-4680(12)00081-8/abstract
15. Hart, A. L. "Modulation of Human Dendritic Cell Phenotype and Function by Probiotic Bacteria." Gut 53.11 (2004): 1602-609. Web. http://gut.bmj.com/content/53/11/1602.long#ref-8
16. Pokusaeva, Karina, Gerald F. Fitzgerald, and Douwe van Sinderen. “Carbohydrate Metabolism in Bifidobacteria.” Genes & Nutrition 6.3 (2011): 285–306. PMC. Web. http://link.springer.com/article/10.1007%2Fs12263-010-0206-6
17. Mayo, Baltasar, and Douwe Van. Sinderen. "Bifidobacteria: Genomics and Molecular Aspects." Norfolk, UK: Caister Academic, 2010. Print.
18. Fallingborg, J. "Intraluminal PH of the Human Gastrointestinal Tract." Danish Medical Bulletin 46.3 (1999): 183-96. PubMed. Web. http://www.ncbi.nlm.nih.gov/pubmed/10421978
19. Azar, Sami T. et al. “Type I (Insulin-Dependent) Diabetes Is a Th1 and Th2-Mediated Autoimmune Disease.” Clinical and Diagnostic Laboratory Immunology 6.3 (1999): 306–310. Web. http://www.ncbi.nlm.nih.gov/pmc/articles/PMC103714/
20. Manicassamy, Santhakumar et al. “TLR2 Dependent Induction of Vitamin A Metabolizing Enzymes in Dendritic Cells Promotes T Regulatory Responses and Inhibits TH-17 Mediated Autoimmunity.” Nature medicine 15.4 (2009): 401–409. PMC. Web. http://www.ncbi.nlm.nih.gov/pubmed/19252500
21. Rook, G A W, and L R Brunet. “Microbes, Immunoregulation, and the Gut.” Gut 54.3 (2005): 317–320. PMC. Web. http://www.ncbi.nlm.nih.gov/pmc/articles/PMC1774411/
22. Rachmilewitz D, Karmeli F, Takabayashi K, et al "Immunostimulatory DNA ameliorates experimental and spontaneous murine colitis." Gastroenterology (2002) 122:1428
23. Hoarau C, Lagaraine C, Martin L, Velge-Roussel F, Lebranchu Y. "Supernatant of Bifidobacterium breve induces dendritic cell maturation, activation, and survival through a Toll-like receptor 2 pathway." J Allergy Clin Immunol. 117 (2006):696–702. Web. http://www.ncbi.nlm.nih.gov/pubmed/16522473
24. Calcinaro, F., Dionisi, S., Marinaro, M., Candeloro, P., Bonato, V., Marzotti, S., … Dotta, F. (2005). "Oral probiotic administration induces interleukin-10 production and prevents spontaneous autoimmune diabetes in the non-obese diabetic mouse." Diabetologia, 48.8 (2005): 1565–1575. Web. http://link.springer.com/article/10.1007/s00125-005-1831-2/fulltext.html
25. Murri, Mora et al. “Gut Microbiota in Children with Type 1 Diabetes Differs from That in Healthy Children: A Case-Control Study.” BMC Medicine 11 (2013): 46. PMC. Web. http://www.ncbi.nlm.nih.gov/pubmed/23433344
26. McCall, Becky. "Probiotics Cut Autoimmunity 33% in Infants at Risk for Diabetes." Medscape. Sep 19, 2014. Web. http://www.medscape.com/viewarticle/831982#vp_1
27. Visser, Jeroen et al. “Tight Junctions, Intestinal Permeability, and Autoimmunity Celiac Disease and Type 1 Diabetes Paradigms.” Annals of the New York Academy of Sciences 1165 (2009): 195–205. PMC. Web. http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2886850/
28. Oriss, Timothy B. et al. "Dynamics of dendritic cell phenotype and interactions with CD4+ T cells in airway inflammation and tolerance." J. Immunol. 174.2 (2005): 854-863. PMC. Web. http://www.ncbi.nlm.nih.gov/pubmed/15634907
Edited by Kathleen Muenzen, a student of Nora Sullivan in BIOL168L (Microbiology) in The Keck Science Department of the Claremont Colleges Spring 2014.
