Ebola Virus NEU2011: Difference between revisions
No edit summary |
m (moved User:YerovO2 to Ebola Virus NEU2011) |
(No difference)
| |
Revision as of 16:53, 22 February 2011
Ebola Virus
Classification
Class: Class V - ssRNA negative-strand viruses
Order: Mononegavirales
Family: Filoviridae
Genus: Ebolavirus
Species: Zaire ebolavirus
[1](based on Baltimore classification system)
Description and Significance
The viral family Filoviridae consists of two distinct genuses, Marburgvirus and Ebolavirus. The family name, from the Latin "filum" meaning "thread" or "filament", describes the threadlike structure of the organisms. Filoviruses are negative-strand RNA viruses, usually between 850 and 920nm in length [2]. In the Ebolavirus genus, four distinct species have been identified, three of which co-circulate in Africa, and are known human pathogens [3].
The Ebola virus (EBOV) strains were first identified in Africa in 1976. Two epidemics occurred almost simultaneously in Zaire and Sudan, killing almost 500 people. In Zaire, an outbreak at a local hospital, possibly due to contaminated hypodermic needles, exhibited an 88% mortality rate for those infected. The predominant symptoms of the virus were hemorrhagic fever and damage to the endothelial cells that form the linings of blood vessels, characterized by severe internal and sometimes external bleeding. The responsible strain was identified and named Ebola, after the Ebola River region where the epidemic occurred. This strain of the virus is referred to as the Zaire virus, and is the type species for the genus [4,5].
Since its discovery over 30 years ago, EBOV research has elucidated much about the mechanism by which an Ebola outbreak occurs. Since the epidemic in Zaire, several more cases have been observed in humans and animals, though no other strain has proved as deadly as that isolated from the Ebola River area [3]. While research has significantly expanded knowledge about how the virus survives between epidemics and how it is passed to human populations, much is still unknown about EBOV. With the modern advent of biological warfare, the contagiousness and lethality of EBOV makes it a potential weapon of bioterrorism [6]. Therefore, further research is important to understand and hopefully identify a vaccine for EBOV, to protect against future epidemics.
Genome Structure
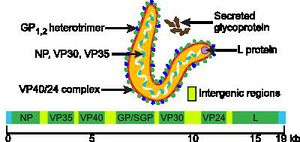
The Ebola genome, as with that of other members of Mononegavirales, is a non-segmented and consists of a single molecule of linear, negative-strand ssRNA. The genome is approximately 19kb long, and constitutes about 1.1% of the viral weight [1, 7, 8].
The Ebola genome has 7 known nucleotide sequences that code for structural and non-structural proteins also known as VPs. The core of the virus is made up of RNA genomic molecules comprised of nucleoprotein (NP) [7].
There are several types of VPs, each with a different function. VP30 proteins play an important role in RNA transcription activation, which is strongly dependent on the concentration of VP30. VP24 the primary matrix protein, and is the most abundant virion component. Its role is unclear. VP35 plays an important role in viral RNA synthesis. It acts as a type of interferon antagonist. VP40 is a matrix protein from the negative strand of RNA. It mainly participates in the assembly of lipid-enveloped viruses by providing a link between the surrounding membrane and the nucleocapsid structure. The protein GP, also known as single surface transmembrane GP, is responsible for attachment and entry into the target cell. The L protein is a non-segmented negative strand RNA, it plays an essential role in catalyzing transcription. The L protein is a RNA-dependent RNA polymerase [1, 7].
All gene expression required for viral replication is carried out within the Ebola virion, with no participation by the host-cell enzymes. Nucleotide sequences at the 3’-terminus are complimentary to similar regions of genetic code in the 5’-end, which does not have a cap. The 3’-end also has no poly(A) trail. Transcription occurs from the 3’-end and results in seven to nine mRNA strands. The Ebola amino acids identity is distinct from that of other Filoviridae viruses, evidenced by its unique method of entry into the host-cell [1, 7].
References
- ICTVdB Management (2006). 01.025.0.02.Ebolavirus. In: ICTVdB - The Universal Virus Database, version 4. Büchen-Osmond, C. (Ed), Columbia University, New York, USA
- Regnery, R.L., K.M. Johnson, and M.P. Kiley, Marburg and Ebola viruses: possible members of a new group of negative-strand viruses. The replication of negative-strand viruses, 1981: p. 971-977.
- Pourrut, X., et al., The natural history of Ebola virus in Africa. Microbes and Infection, 2005. 7: p. 1005-1014.
- McCormick, J.B., et al., Biologic Differences between Strains of Ebola Virus from Zaire and Sudan. The Journal of Infectious Diseases, 1983. 147(2): p. 264-267.
- Heymann, D.L., et al., Ebola Hemorrhagic Fever: Tandala, Zaire, 1977-1978. The Journal of Infectious Diseases, 1980. 142(3): p. 372-376.
- Groseth, A., et al., Hemorrhagic Fever Viruses as Biological Weapons, in Bioterrorism and Infectious Agents: A New Dilemma for the 21st Century, I.W. Fong and K. Alibek, Editors. 2009, Springer New York. p. 169-191.
- Ascenz, P., et al. "Ebolavirus and Marburgvirus: Insight the Filoviridae Family." Ebolavirus and Marburgvirus: Insight the Filoviridae Family. 2008. 29(3): p. 151-85.
- Paustian. "19-20 Ebola Is a Filamentous Virus with a Single-stranded RNA Genome." The Microbial World :: A Look at All Things Small. 08 Nov. 2008. Web. 21 Feb. 2011. link
Images a, b courtesy of Center for Biological Counterterrorism and Emerging Diseases CBC-ED image gallery
Image c courtesy of The Microbial World