Effects of Scleroderma on Human Microbiome
Introduction
Scleroderma (or systemic sclerosis, SSc) is an autoimmune rheumatological disorder in which hardening of the skin and other organs occurs through increased amounts of collagen in the skin. [1] Additionally, there are significant impacts on the vascular system by this hardening. There are two types of scleroderma, localized and systemic. Localized is when the hardening occurs exclusively in the skin and muscular tissues. Systemic occurs when the hardening is widespread throughout many organs in the body. Additionally, there are a variety of factors suggested in the pathogenesis of scleroderma, namely, environmental, genetic, and oxidative stressors. Further, sex is known to be a major risk factor for scleroderma and autoimmune disorders in general, with autoimmune disorders occurring in those of the female sex at double the frequency. [2] A potential cause of these sex disparities is the difference in the human microbiome between the sexes. It has been noted that autoimmunity is able to be characterized by changes in the gut microbiome. Changes in the gut microbiome occur more frequently in the female sex.[3] Furthermore, genetic factors such as incomplete X-inactivation contribute to a higher prevalence of autoimmunity in the female sex.[4]
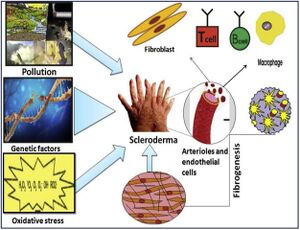
Microbiome
First, it should be noted that perhaps the most overt connection to microbes in SSc is that the leading cause of death in SSc patients is Interstitial lung disease (ILD).[5] More specifically, classes of pneumonia within ILD can be caused by various microbes, including Streptococcus pneumoniae.[6]
It has been observed specifically in the GI tract that scleroderma symptoms are associated with a change in the gut microbiome, including decreases in microbial diversity. [7]In studies of scleroderma patients’ gut microbiomes who displayed the SSc GI tract phenotype (scleroderma patients with GI symptoms), increased levels of Fusobacterium and Prevotella were observed.[8] Such a shift in the gut microbiome suggests that the gut microbiome contributes to the development of the SSc GI tract phenotype. Moreover, due to the established link between Prevotella in the gut and diet, it is suggested that dietary changes to lower the levels of Prevotella in the gut microbiome might reduce the effects of the SSc GI tract phenotype.
Differences in microbial diversity are typical of various forms of scleroderma.[7] Notably, when comparing the microbiomes of patients with diffuse cutaneous systemic sclerosis (DcSSc) and microbiomes of patients with limited cutaneous systemic sclerosis (LcSSc), patients with DcSSc had lowered abundance of Lactobacillus and Coprococcus and raised abundance of Paludibacter. Furthermore, less Coproccus is seen in patients with nonalcoholic fatty liver disease, specifically in the metabolic pathway leading to liver fibrosis. This knowledge of the Coproccus-liver fibrosis metabolic pathway link has led researchers to suggest further research into the potential role of Coproccus in the development of fibrosis phenotypes.
Changes in microbial diversity have not only been associated with scleroderma but other autoimmune disorders as well. Such disorders include inflammatory bowel disease, rheumatoid arthritis, and systemic lupus erythematosus, among others.
Genetics
Gene Expression
Alistipes, Bacteroides, and butyrate-producing bacteria are less abundant in SSc patients than healthy controls; Clostridium and Streptococcus are more abundant.[9] Perhaps more importantly, Eggerthella lenta and Clostridium boltae are also more abundant in SSc patients. E. lenta, specifically, plays a potentially critical role in upregulating Th17-activating genes in SSc patients. T helper 17 (Th17) cells are pro-inflammatory and thus potential contributors to the phenotype of organ inflammation in SSc. By upregulating Th17-activating genes, inflammation observed in SSc patients might be induced or worsened.
Taxonomy
In SSc patients, there is a shift at the phylum level of microbiome composition.[7] More specifically, the ratio of Firmicutes to Bacteroidetes increases -- meaning that the abundance of Bacteroidetes decreases -- a trend shown in patients with obesity. Such a change in relative abundance suggests a potential mechanism in SSc patients struggling to maintain a healthy weight. The suggestion of gut microbiome changes leading to weight management difficulties is supported by research that has indicated microbiome changes are associated with the development of digestive dysbiosis, a condition characterized by diverse possible GI symptoms, including obesity.[10] Furthermore, there have been other changes in the gut microbiome characteristic of various GI disorders such as IBD. Some of these changes are increases in the abundance of Actinobacteria, Fusobacteria, and Verrucomicrobia.[7]
Genus level differences were also noticed between Scleroderma patients and healthy controls. For example, Bacteroides have been observed as less abundant in SSc patients. The genus Bacteroides helps to protect from mucosal inflammation and pathogenic bacteria. Moreover, decreases in Bacteroides have been seen in immunodeficiency disorders such as Crohn’s Disease. Additionally, butyrate-producing bacteria species which facilitate healthy gut function decrease in abundance in SSc patients, namely, Clostridium and Faecilibacterium. Moreover, butyrate-producing bacteria help to protect from mucosal inflammation as well, furthering the issue of mucosal inflammation resulting from taxonomic shifts in the gut microbiome of SSc patients.
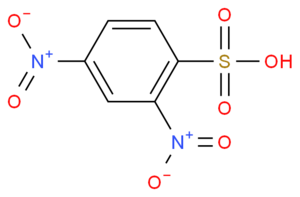
Furthermore, there have been links between changes in the metabolome of SSc patients and healthy controls.[11] Increases in the Desulfovibrio genus in the gut microbiome have been associated with increased levels of the alpha-N-phenylacetyl-l-glutamine and 2,4-dinitrobenzenesulfonic acid metabolites. The effect of increased plasma levels of these metabolites is currently unclear, though it has been suggested that they might contribute to the development or progression of SSc.
Sex-Linked Pathogenesis
Beyond the previously mentioned potential microbial reasons for the female sex being more likely to develop autoimmune disorders, the presence of two X-chromosomes has been long documented as a genetic factor in the development of autoimmunity.[4] Approximately 15% of X-linked genes are not deactivated during X-inactivation. This can lead to overexpression of immune genes in certain individuals that manifest in an autoimmune phenotype.
Sex-linked disparities in autoimmunity frequency have also been shown to result from an increased innate and adaptive immune response to pathogens in the female sex.[12] Moreover, mouse studies have shown that female-sex mice were more likely to over-respond to an antigen than male-sex mice, resulting in female-sex mice experiencing autoimmunity to some degree.
Conclusion
Systemic sclerosis is a rheumatological autoimmune disorder characterized by the hardening of organs in the body. Associated with SSc are diverse symptoms and phenotypes, many of which have been associated with changes in the microbiome of SSc patients. More specifically, various phenotypes, such as the GI tract phenotype and digestive dysbiosis, are specifically linked to changes in microbiome composition. These microbiome changes have been documented with large phyla and genus shifts, not to mention abundance and relative abundance changes. Moreover, these shifts have resulted in non-GI symptoms. There are elements of immunosuppression and exacerbation of mucosal inflammation that result from changes in the microbiome. Even further, the bacterium E. lenta has been shown to alter the expression of Th17-activating genes and induce inflammation as well. The contribution of the microbiome to the phenotypical presentation of scleroderma is well established. In addition, various genetic factors have been identified, including sex-linked risk factors. Yet, microbiologists continue to explore the contribution of the microbiome to scleroderma for potential treatments, such as reestablishing a typical microbiome, a proposed treatment for some GI symptoms. Therefore, the field of knowledge on the mechanisms of how the microbiome impacts scleroderma is constantly evolving.
References
- ↑ Singh, Deependra, Arun KS Parihar, Satish Patel, Shikha Srivastava, Prakriti Diwan, and Manju R. Singh. 2019. “Scleroderma: An Insight into Causes, Pathogenesis and Treatment Strategies.” Pathophysiology 26 (2): 103–14.
- ↑ Angum, Fariha, Tahir Khan, Jasndeep Kaler, Lena Siddiqui, and Azhar Hussain. 2020. “The Prevalence of Autoimmune Disorders in Women: A Narrative Review.” Cureus, May.
- ↑ Gomez, Andres, David Luckey, and Veena Taneja. 2015. “The Gut Microbiome in Autoimmunity: Sex Matters.” Clinical Immunology 159 (2): 154–62.
- ↑ 4.0 4.1 Carrel L. and Willard H. 2005 “X-Inactivation Profile Reveals Extensive Variability in X-Linked Gene Expression in Females.” Nature.
- ↑ Domiciano, Diogo S., Eloisa Bonfá, Claudia T.L. Borges, Ronaldo A. Kairalla, Vera L. Capelozzi, Edwin Parra, and Romy Beatriz Christmann. 2011. “A Long-Term Prospective Randomized Controlled Study of Non-Specific Interstitial Pneumonia (NSIP) Treatment in Scleroderma.” Clinical Rheumatology 30 (2): 223–29.
- ↑ “Respiratory Syncytial Virus (RSV) Infection Rhinovirus Infection.” n.d.
- ↑ 7.0 7.1 7.2 7.3 Tan, Tze Chin, Maria Noviani, Ying Ying Leung, and Andrea Hsiu Ling Low. 2021. “The Microbiome and Systemic Sclerosis: A Review of Current Evidence.” Best Practice and Research: Clinical Rheumatology. Bailliere Tindall Ltd.
- ↑ Volkmann, Elizabeth R., Yu Ling Chang, Nashla Barroso, Daniel E. Furst, Philip J. Clements, Alan H. Gorn, Bennett E. Roth, et al. 2016. “Association of Systemic Sclerosis with a Unique Colonic Microbial Consortium.” Arthritis and Rheumatology 68 (6): 1483–92.
- ↑ Plichta, Damian R., Juhi Somani, Matthieu Pichaud, Zachary S. Wallace, Ana D. Fernandes, Cory A. Perugino, Harri Lähdesmäki, et al. 2021. “Congruent Microbiome Signatures in Fibrosis-Prone Autoimmune Diseases: IgG4-Related Disease and Systemic Sclerosis.” Genome Medicine 13 (1).
- ↑ Bratoiu, Ioana, Alexandra Burlui, Patricia Richter, Anca Cardoneanu, Ciprian Rezus, and Elena Rezus. 2021. “Digestive Dysbiosis in Systemic Scleroderma: A Review.” Journal of Interdisciplinary Medicine 6 (2): 53–59.
- ↑ Bellocchi, Chiara, Álvaro Fernández-Ochoa, Gaia Montanelli, Barbara Vigone, Alessandro Santaniello, Christian Milani, Rosa Quirantes-Piné, et al. 2018. “Microbial and Metabolic Multi-Omic Correlations in Systemic Sclerosis Patients.” Annals of the New York Academy of Sciences 1421 (1): 97–109.
- ↑ Taneja, V., Behrens, M., Mangalam, A., Griffiths, M. M., Luthra, H. S., & David, C. S. (2007). New humanized HLA–DR4–transgenic mice that mimic the sex bias of rheumatoid arthritis. Arthritis & Rheumatism, 56(1), 69-78.
Edited by Andrew Van Horn, student of Joan Slonczewski for BIOL 116 Information in Living Systems, 2021, Kenyon College.
