Enterococcus faecalis: Difference between revisions
| Line 17: | Line 17: | ||
==Description and significance== | ==Description and significance== | ||
Enterococcus are gram-positive cocci that can survive harsh conditions in nature | Enterococcus are gram-positive cocci that can survive harsh conditions in nature. They can be found in soil, water, and plants. They are also known to colonize the gastrointestinal and gential tracts of humans. While there are over 15 species of the Enterococcus genus, 80-90% of clinical isolates are E. faecalis. | ||
In the last decade, vancomycin-resistnat enerococci | In the last decade, vancomycin-resistnat enerococci have become a major challenge. In general, entercoccal isolates with lowered susceptibility to vancomycin can be categorized as vanA, vanB, and vanC. vanA and vanB pose the greatest threat because they are the most resistant and the resistance genes are carried on a plasmid. Since the resistance genes are carried on a plasmid they readily transferable. E. faecalis strains are categorized as vanA isolates. E. faecalis are also resistant to teicoplanin. Enterococcal strains that are vancomycin dependent have been reported, but are rare and are less common that vancomycin-resistant strains. | ||
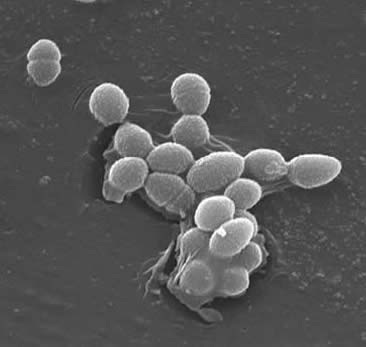
Enterocci typically form short chains or are arranged in pairs | Enterocci typically form short chains or are arranged in pairs. However, under certain growth conditions, they elongate and appear coocobacillary. In general, enterococci are alpha-hemlytic. Some possess the group D Lancefield antigen and can be detected using monoclonal antibody-based agglutination tests. Enterococci are typically catalase negative, grow over a wide temperature range from 10 to 42°C, and are anaerobic. They are able to grow in 6.5% NaCl, can hydrolyze esculin in the presence of 40% bile salts and are pyrrolidonyl arylamidase and leucine arylamidase positive. | ||
==Genome structure== | ==Genome structure== | ||
Revision as of 22:27, 29 April 2007
Classification
Higher order taxa
Bacteria; Firmicutes; Bacilli; Lactobacillales; Enterococcaceae; Enterococcus
Genus
Enterococcus faecalis
Description and significance
Enterococcus are gram-positive cocci that can survive harsh conditions in nature. They can be found in soil, water, and plants. They are also known to colonize the gastrointestinal and gential tracts of humans. While there are over 15 species of the Enterococcus genus, 80-90% of clinical isolates are E. faecalis. In the last decade, vancomycin-resistnat enerococci have become a major challenge. In general, entercoccal isolates with lowered susceptibility to vancomycin can be categorized as vanA, vanB, and vanC. vanA and vanB pose the greatest threat because they are the most resistant and the resistance genes are carried on a plasmid. Since the resistance genes are carried on a plasmid they readily transferable. E. faecalis strains are categorized as vanA isolates. E. faecalis are also resistant to teicoplanin. Enterococcal strains that are vancomycin dependent have been reported, but are rare and are less common that vancomycin-resistant strains. Enterocci typically form short chains or are arranged in pairs. However, under certain growth conditions, they elongate and appear coocobacillary. In general, enterococci are alpha-hemlytic. Some possess the group D Lancefield antigen and can be detected using monoclonal antibody-based agglutination tests. Enterococci are typically catalase negative, grow over a wide temperature range from 10 to 42°C, and are anaerobic. They are able to grow in 6.5% NaCl, can hydrolyze esculin in the presence of 40% bile salts and are pyrrolidonyl arylamidase and leucine arylamidase positive.
Genome structure
Describe the size and content of the genome. How many chromosomes? Circular or linear? Other interesting features? What is known about its sequence? Does it have any plasmids? Are they important to the organism's lifestyle?
Cell structure and metabolism
Describe any interesting features and/or cell structures; how it gains energy; what important molecules it produces.
Ecology
Describe any interactions with other organisms (included eukaryotes), contributions to the environment, effect on environment, etc.
Pathology
How does this organism cause disease? Human, animal, plant hosts? Virulence factors, as well as patient symptoms.
Application to Biotechnology
Does this organism produce any useful compounds or enzymes? What are they and how are they used?
Current Research
Enter summaries of the most recent research here--at least three required
References
example:
De la Maza, Luis M., Marie T. Pezzlo, and Janet T. Shigei. Color Atlas of Medical Bacteriology. Washington, DC: American Society for Microbiology, 2004.
Edited by Richard A. Martinez of UC San Diego, student of Rachel Larsen.