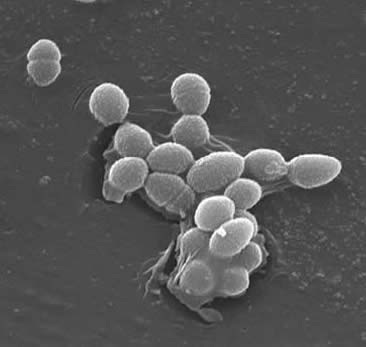
Enterococcus faecalis
Classification
Higher order taxa
Bacteria; Firmicutes; Bacilli; Lactobacillales; Enterococcaceae; Enterococcus
Species
Enterococcus faecalis
Description and significance
Enterococci are Gram-positive cocci that can survive harsh conditions in nature. They can be found in soil, water, and plants. Some strains are used in the manufacture of foods whereas others are the cause of serious human and animal infections (e.g. they are known to colonize the gastrointestinal and genital tracts of humans). They are associated with both community and hospital acquired infections. Enterococci can grow at a temperature range of 10 to 42°C and in environments with broad pH values. Some are known to be motile. While there are over 15 species of the Enterococcus genus, 80-90% of clinical isolates are E. faecalis (8). Enterococci typically form short chains or are arranged in pairs. However, under certain growth conditions, they elongate and appear cocobacillary. In general,Enterococci are alpha-hemolytic. Some possess the group D Lancefield antigen and can be detected using monoclonal antibody-based agglutination tests.Enterococci are typically catalase negative, and are anaerobic. They are able to grow in 6.5% NaCl, can hydrolyze esculin in the presence of 40% bile salts and are pyrrolidonyl arylamidase and leucine arylamidase positive (8). Enterococci have proven to present a therapeutic challenge because of their resistance to many antimicrobial drugs, “including cell-wall active agents; aminoglycosides, penicillin and ampicillin, and vancomycin” (1). The Enterococci have the capacity to acquire a wide variety of antimicrobial resistance factors, which present serious problems in the management of patients with enterococcal infections (8). In general, enterococcal isolates with lowered susceptibility to vancomycin can be categorized as vanA, vanB, and vanC. vanA and vanB pose the greatest threat because they are the most resistant and the resistance genes are carried on a plasmid. Since the resistance genes are carried on a plasmid they are readily transferable, E. faecalis can transfer these plasmids by conjugation (2). E. faecalis are also resistant to teicoplanin. Enterococcal strains that are vancomycin-dependent have been found, but are rare and less common than vancomycin-resistant strains (referred to as “vancomycin-resistant Enterococci” or “VRE”) (1).
Genome structure
Due to many public health dangers, the genome sequence data from a strain of Enterococcus was necessary. The strain chosen for genome DNA sequencing was E. faecalis V583, the first vancomycin-resistant isolate in the United States. The genome of strain V583 was sequenced by The Institute for Genome Research (TIGR). The enterococcal genome shows E. faecalis is metabolically diverse and contains a wide range of regulatory systems. Strain V583 contains four DNA molecules: the main 3,218,030 base pair bacterial chromosome and three circular plasmids. The chromosome contains about 3,500 open reading frames (ORFs), about 1/3 of these ORFs have no assignable function (8). The three plasmids are circular DNA molecules identified as Plasmid-1, Plasmid-2, and Plasmid-3. Plasmid-1 contains 66,320bp, Plasmid-2 contains 17,963bp, and Plasmid-3 contains 57,660bp. The plasmids encode a number of genes, including, transposases, multi-drug resistance proteins, and a ppGpp-regulated growth inhibitor (8). The average G+C composition of the E. faecalis chromosome is 37.38%. Since the DNA molecule is so large, regional deviations from the average occur. One of these locations is the large segment associated with the “vancomycin resistance gene cluster positioned near 2.22 Mb,” showing a large increase in percent G+C content (8). These differences associated with antibiotic resistance or virulence suggested the acquisition of genetic material from a foreign species through horizontal transfer. It is still unknown whether the transfers are responsible for the variations in DNA makeup. The information contained in the genome of E. faecalis V583 will greatly aid the understanding of how the organism has adapted to be a versatile human pathogen. Using comparative genomics, the role of the different regulatory elements will be better understood in how they respond to various environmental stresses and in the expression of potential virulence factors (8). More studies like these will suggest new medicines to bacterial infections caused by the Enterococci. The genome also contains 3 Ebp (encoding for the endocarditis biofilm-associated pili) operons which are important for biofilm production of E. faecalis strain OG1RF. This strain uses these operons to produce surface pili. . The surface pili are used for “attachment to the host surfaces and are antigenic in humans during endocarditis” (9).
The TIGR contains a complete list of genes for the E. faecalis chromosomes.
Cell structure and metabolism
Cell Metabolism
The Enterococci inhabit harsh environments, like the intestinal tracts of humans and animals. Growth under these hostile conditions requires that E. faecalis have a metabolism that is flexible. E. faecalis are capable of not only fermentation to produce lactic acid but also can “catabolize a spectrum of energy sources from carbohydrates, glycerol, lactate, malate, citrate, diamino acids and manyα-keto acids” (3). It has been shown that under selected growth conditions E. faecalis can enhance growth through oxidative phosphorylation using a proton motive force established by electron transport. A consequence of “nascent respiration is production of potent oxidants” (e.g. superoxide and hydrogen peroxide), oxidative stress the E. faecalis can tolerate (3). The tolerance of this stress, combined with other severe growth conditions, allows the E. faecalis to grow at 10 to 45°C, in bile salts, and at extremely low and high pHs. In addition, E. faecalis can resist azide, detergents, heavy metals, and ethanol. Since E. faecalis can utilize varied sugar sources it can live in diverse environments, especially in the intestine where nutrients are limited (2). In the intestine, E. faecalis derive most of their energy from the fermentation of non-absorbed sugars. E. faecalis can also get energy by degrading mucins, a carbohydrate that is heavily glycosylated and produced by intestinal goblet cells (2). The E. faecalis uses a “phosphoenolypyruvate phosphotransferase system (PTS) to sense sugars outside the cell and couples uptake of sugars with phosphorylation” (2). In doing so, less energy (ATP) is wasted compared to how sugar is accumulated by non-PTS systems. Sugars metabolized by E. faecalis are include: D-glucose, D-fructose, lactose, maltose (all PTS substrates). In Enterococci, PTS also regulates glycerol metabolism, inducer expulsion, and catabolite repression (8). E. faecalis is one of a few low-G+C content Gram-positive bacteria that expel sugar during growth on glucose, a phenomenon known as inducer expulsion (8). E. faecalis can even ferment glycerol under aerobic and microaerophillic conditions. E. faecalis can also grow on glycerol under anaerobic conditions because it “expresses a gene for NAD+-linked anaerobic glycerol dehydrogenase activity” (8). Glycerol can cross the cell membrane using a protein called the glycerol diffusion facilitator (GlpF). The GlpF makes the concentration of glycerol inside and outside the cell equal, the protein is inhibited by glycolysis. E. faecalis are able to live in extreme alkaline pH and high salt concentration. These traits require cation transport to maintain the constant cytosolic ion composition essential for homeostasis. All cells must expel excess sodium to maintain cytosolic concentrations in range that favors homeostasis. E. faecalis expresses both a Na+/H+ antiporter and a vacuolar-type ATPase (2). Potassium is a major intracellular cation. The potassium concentration within E. faecalis of 0.4 to 0.6 M is essential for normal cellular metabolism, it “neutralizes intracellular anions, activates diverse enzymes, and regulates cytosolic pH” (8). E. faecalis expresses at least two potassium transporters to perform the regulation, KtrI and KtrII along with the Kep system for K+ extrusion (8). Although it is known that KtrI and KtrII are K+ uptake systems (they are K+/H+ symporters), little more is known the proteins.
Cell Structure Enterococci are Gram-positive cocci that typically form short chains or are arranged in pairs (3). Under certain growth conditions they can elongate and appear coccobacillary. The cell wall of E. faecalis is 20 to 38% of the dry cell weight (in the exponential and stationary phase cells). Given that the E. faecalis is a Gram-positive bacterium there are three main components that make up its cell wall: peptidoglycan, teichoic acid, and polysaccharide. 40% of the cell wall is made up of peptidoglycan, while the rest of the cell wall is made up of a “rhamnose-containing polysaccharide and a ribitol-containing teichoic acid” (3). The peptidioglycan functions (as in most Gram-positive cells) to resist bursting induced by high cytoplasmic osmotic pressure. E. faecalis is generally considered a non-encapsulated organism, shown by the “lack of a detectable mucoid phenotype” (3). However, subsets of E. faecalis isolates possess a capsular polysaccharide. E. faecalis can exchange genetic material (plasmids) by conjugation processes induced by small peptide pheromones (2). Surface protein “aggregation substances that recognize a specific ligand on recipient cells” ensure successful connections for conjugation (2). E. faecalis also have the capability to make surface pili which can lead to the formation of a biofilm. The E. faecalis strains that cause endocarditis contain large amounts of these pili. The pili allow for attachment to host surfaces (e.g. the heart tissue). The strains of E. faecalis that cause endocarditis produce the “biofilm significantly more often and also to a greater degree than non-endocarditis isolates” (9).
Ecology
E. faecalis is a very diverse species of enterococci. It interacts with many other organisms and has effects on the environment.
The enterococci are members of the bacterial community inhabiting the large bowel in humans. They also are a natural part of the intestinal flora in most other mammals and birds. The enterococci are also found in soil, on plants, and in water. When they are found in water it is typically because the water had been contaminated with fecal matter. Although E. faecalis do not appear too often in plants, other strains of Enterococci have been found among plants. This is most likely do to a contaminated water source (10).
The ecology of antibiotic resistance and virulence gene transfer in the environment is still not well understood. Insects, such as houseflies (HF), that develop in decaying organic material may transmit antibiotic-resistant bacteria from the manure of animals and other decaying organic substrates to residential settings. The habitats in which the HF develops (e.g. manure), its dependence on a live microbial community, its feeding mechanism (regurgitation), its attraction to human food, and its ability to fly long distances make this insect a very good candidate for spreading fecal bacteria, including human and animal pathogens, and possibly antibiotic-resistant strains of enterococci (9).
The enterococci also pose problems from the food safety perspective. Enterococci have previously been isolated from milk, cheese, meats, and from raw produce. A study was able to isolate, identify, and screen for antibiotic resistance and virulence genes in enterococci from HF in fast-food restaurants in Kanasas. This study showed that houseflies in food-handling and serving facilities can carry antibiotic-resistant and potentially virulent enterococci that have the capacity for horizontal transfer of antibiotic resistance genes to other bacteria (9).
The effects that E. faecalis has on the environment tend to be more negative. They typically contaminate water supplies which can lead to infected plants as well as infections in people. The antibiotic factors can also be transported by various insects and animals, leading to increasing numbers of infectious virulent E. faecalis.
Describe any interactions with other organisms (included eukaryotes), contributions to the environment, effect on environment, etc.
Pathology
Enterococci have emerged in the last part of the 20th century as a major cause of nosocomial infections, and within this group Enterococcus faecalis causes the majority of human enterococcal infections. These infections may be local or systematic and include uninary tract and abdominal infections, wound infections, bacteremia, and endocarditis. Enterococci are part of the many bacteria that make up the normal human intestinal tract and are capable of surviving numerous environmental challenges such as temperature extremes and the presence of bile salts. Their acquisition of resistance to multiple antibiotics has made these bacteria a major health problem. The National Nosocomial Infection Surveillance (NNIS) system has reported a rapid rise in the incidence of infection and colonization with vancomycin-resistant enterococci (VRE) since 1989. This can mean serious health problems, which include the lack of available antimicrobial therapy for VRE infections, because most VRE strains harbor resistance to multiple antibiotics (e.g. aminoglyscoides and ampicillin) and “vancomycin is often the antibiotic of last resort”.
The transfer of vanocmycin resistant genes from VRE to other gram-positive pathogens is a serious public health concern. The most common way that the E. faecalis cause infection occurs in a hospital/long term care facility is transmission of E. faecalis between patients. Enterococci can be carried on the hands of health care workers and be carried (transferred) from one patient to another. It has been shown that VRE inoculated on the hands can persist for up to 60mins. The transmission from a health care worker’s hands to the patient could take place direct contact with the patient’s intravenous or urinary catheters. Rectal thermometers, not properly cleaned after use, can transmit the VRE from patient to patient as well. Sometimes the transmission can result in colonization of the patient’s GI tract with the acquired strain. The new strain then becomes part of the patient’s endogenous flora. The acquired strain, carrying antibiotic resistance genes, is able to live in the GI trtact. Infections then arise from these newly acquired E. faecalis strains.
E. faecalis can cause many bodily infections. The most common infection caused by enterococci is infection of the urinary tract. E. faecalis can cause lower urinary tract infections (UTI), such as cystisis, prostatitis, and epididymitis. E. faecalis are also found in intra-abdominal, pelvic, and soft tissue infections. The E. faecalis are the third leading cauase of nosocomial bacteremia. The source of bacteremia is most often the urinary tract, occurring from an infected intravenous catheter (for example). Endocarditis is the most serious enterococcal infection. Due to the intrinsic resesitence of enterococci to many antibiotics, treatment of endocarditis is difficult. In many cases of endocarditis, antibiotic treatment fails and surgery to remove the infected valve is necessary. Less common infectrions caused by E. faecalis include meningitis, hematogenous, osteomyelitis, septic arthritis, and (very rare) pneumonia.
Application to Biotechnology
Enterococci have been studied for possible use as a probiotic, a dietary supplement that contains living non-virulent microbial cells that when ingestd are thought to beneficially affect the composition of the intestinal microflora. Administration of the E. faecalis strain has been reported to reduce diarrhea. The E. faecalis probiotics are used in chicken/pig/cattle feed to lower diarrhea in these animals (10).
Due to the high disease causing properties of E. facalis, much more research has been conducted on how to stop the virulence of E. facalis than the benefical use of E. facalis.
Current Research
1.) Hope for infections caused by Enterococcus faecalis (6)
It can be seen from my article above on E. faecalis that vancomyacin resistance bacteria pose many problems to society, causing bacterial infections in humans that are difficult to treat. The enterococcal infections are challenging because the organisms have the ability to quickly acquire and disseminate resistance genes. Ceftobiprole (BPR) was used as an investigational cephalosporin against gram-positive cocci. BPR is a broad-specturm parenteral cephalosporin with high affinities from gram-positive and gram-negative penicillin-binding proteins. It also shows stability against hydrolysis by B-lactamases.
This study examined the activity of BRP against a large collection of E. faecalis, looking at BPR bactericidal activity against vancomycin-resistant isolates. The study found that susceptibly to BPR in E. faecalis is not affected by the presence of vancomycin resistance or by B-lactamase production in enterococci. In the strains of E. faecalis showing either vancomycin resistance (VanA and VanB phenotypes) or ampiciliin resistance, the BPR was bactericidal. The researchers suggested that their results supported the fact that BPR would likely exhibit bactericidal activity against E.faecalis at a dose of 750mg. This does inhibited 100% of their E.faecalis activity. Although the presence of the enzyme B-lactamase is rare in E. faecalis, its presence compromises the use of the most effective antienterococcal B-lactams (ampicillin). BPR is a poor substrate for B-lactamase enzyme, explaining its excellent activity against B-lactamase-producing E. faecalis.
This research demonstrated that BPR has potent activity against a very large collection of E. faecalis isolates. The activity of the BPR was not affected by vancomycin resistance or production of ß-lactamase. Therefore, BPR is a promising agent for use to treat B-lactamase producing and vancomycin-resistant E. faecalis infections in humans.
2.) Examining howEnterococcus faecalis become resistant to the drug linezolid (7)
When antibiotics are used in the treatment of a bacterial infection, they can have an impact on the intestinal flora. Resistant bacteria can be selected during treatment, such as the enterococci, and are potentially pathogenic. The dynamics of the emergence of resistance are an issue for new antibiotics, because such emergence could risk the usefulness and life span of antibiotics.
This study examined the emergence of resistance to antibiotics by E. faecalis. More specifically, the researchers looked at resistance to linezolid (the first of a new class of antibiotics, the oxazolidinones), modeling the effect of different regimens of linezolid on Enterococcus faecalis in gnotobiotic mice. Linezolid can be used against multiple-drug-resistant gram-positive cocci, including VRE. It inhibits bacterial protein synthesis by binding specifically to a domain in the 50S ribosomal subunit and is not affected by the resistance mechanisms that affect other antibiotics. This study looked at the rate of emergence of linezolid-resistant E. faecalis mutants in the digestive tract of the mice. The does of linezolid was fed in water with a doses varying from 0.5, 0.005 or 0.005 g/L. The mutants were all dependent on the linezolid regimen given, levels of resistance increased with the duration of exposure. No mutants were isolated in the absence of linezolid, suggesting that de novo resistance to linezolid was uncommon in the enterococci.
The research found that a mutation in a single 23S rRNA gene is the critical step in the emergence of linezolid resistance. Primary colonization with single-mutation mutants were observed as early as 5 days after treatment initiation in mice.
These experiments involving mice help explain the pattern of emergence of resistance to linezolid observed in clinical isolates. The research team showed that dose is critical to the dynamics of resistance. These findings should help define the best therapeutic strategies to minimize the emergence of resistance in the clinical setting, possibly saving many lives.
3.) A Ser/Thr kinase in Enterococcus faecalis is found to mediate antimicrobial resistance (8)
Enterococcus faecalis causes many of the antibiotic resistant infections in hospitals, a consequence of its inherent resistance to certain antibiotics and of its ability to survive and proliferate in the intestinal tract.
The genetic basis of the resistance seen in E. faecalis is not well known, this research attempts to identify a cause of the resistance. The researchers show that PrkC, a one-component signaling protein containing a eukaryotic-type Ser/Thr kinase domain, allows for inherent antimicrobial resistance and intestinal persistence of E. faecalis. They found that an E. faecalis mutant lacking PrkC grew at a wild-type rate in the absence of antimicrobial stress but exhibited enhanced sensitivity to cell-envelope-active compounds, including antibiotics that targeted cell-wall biogenesis and bile detergents.
PrkC regulates physiological processes in E. faecalis that are key to its success as a nosocomial pathogen. The researchers’ predicted stucture of PrkC was a cytoplasmic kinase domain separated by a transmembrane segment from extracellular domains thought to bind uncross-linked peptidoglycan. This suggested that PrkC was a transmembrane receptor that monitors the integrity of the E. faecalis cell wall and mediates adaptive responses to maintain cell-wall integrity.
PrKC is important for E. faecalis to cause nosocomial infections, suggesting that the signaling protein is a target for the development of therapies to prevent infections by antibiotic-resistant enterococci.
References
NOTE: The numbers above serve as a note to which reference below was used for that paragraph.
4.) De la Maza, Luis M., Marie T. Pezzlo, and Janet T. Shigei. Color Atlas of Medical Bacteriology. Washington, DC: American Society for Microbiology Press, 2004.
10.) Gilmore, Michael. The Enterococci: Pathogenesis, Molecular Biology, and Antibiotic Resistance. Washington, DC: American Society for Microbiology Press, 2002.
Edited by Richard A. Martinez of UC San Diego, student of Rachel Larsen.