Evolution of HIV
Human immunodeficiency virus/acquired immune deficiency syndrome (HIV/AIDS) is a global problem of a staggering magnitude that not only impacts human health but economic and social development. The human immunodeficiency virus, HIV, is a lentivirus that depletes the body of CD4+ T helper cells, which are critical for regulating the immune response. A weakened immune system then leads to AIDS, defined as a CD4+ count of lower than 200 cells per milliliter of blood, where the patient is unable to fight opportunistic infections. Currently, there is no cure for the virus. In 2013, it was estimated that 35 million people were living with HIV, with over 2 million new infections that year1. It is transmitted in many ways, such as unprotected sex, infected blood donations, intravenous drug use, and from mother to child. In order to understand the lethality of HIV and how it evades current treatment options despite continued efforts to fight this disease, it is critical to examine the molecular components of this virus and its history.
Early Beginnings

HIV Subtypes
What people think of HIV today is actually comprised of several strains. HIV-1 is the predominant strain that has spread worldwide, while HIV-2 is still confined to parts of Africa. Furthermore, HIV-1 is divided into several subtypes, of which type M is the most predominant globally, with subtypes N, O, and P being much rarer. As well, HIV-1 type M is further subdivided into several subclades.
HIV is descended from a virus that infects African primates, called simian immunodeficiency virus (SIV)219. This was not a one time thing. For instance, HIV-2 jumped from sooty mangabeys to humans many times to create a different strain each time a unique infection was established, which was confirmed by viral sequence analysis that showed both extensive diversity and common ancestry18. However, most of the time, SIV is too weak to cause AIDS in humans. It requires mutations and adaptations to the human host, and it has been suggested that it needs to be transmitted between multiple human hosts in a short time frame for SIV to accumulate useful mutations that would change it into HIV. This can be seen in subtypes of HIV that are not globally relevant. They are much weaker pathogens, replicate and are passed on poorly, and are usually locally constrained 3. This indicates that the main subtypes of HIV that are globally important have accumulated many mutations and were shuttled around quickly to cause the global health crisis today. As they were passed around, they mutated even further and adapted to the populations of the places they are in (Figure 1).
Using evolutionary analysis, HIV-1 subtype M has been dated to around 1920. This was done with early sequence data of HIV viruses, as well as collection of samples from blood and tissue that was then used to create phylogenetic trees of HIV. The data suggests that HIV subtype M started in the Democratic Republic of the Congo, in an area now known as Kinshasa. Both a high density of samples and a high diversity of sequence from samples in Kinshasa have supported this. From there, HIV migrated outwards to surrounding areas, helped by transport links such as river traffic and railway connections that were newly created4.
The Start of a Pandemic
Around 1960, the evolution and growth of HIV exploded and transmission increased exponentially. This can be attributed to the establishment of reservoirs of infection in sex workers, as well as the use of unsterilized needles for injections in medical clinics in vaccine drives. The disease was then spread to Haiti, where it made its way to the United States. This is all in contrast to subtype O, which never made it out of the region. This indicates that all the right spatial, temporal, and societal variables converged to give HIV the boost it needed to become a pandemic4.
In the United States, 1981, doctors first noticed the emergence of two rare diseases that tended to only afflict the elderly and the immunocompromised, Pneumocystis pneumonia and Kaposi’s sarcoma. What was startling about this outbreak was the fact that these disease were found in young and otherwise healthy adults. These opportunistic infections in seemingly healthy adults alerted the CDC and the American public to a previously unknown disease with an unknown means of transmission. Public anxiety grew, and it became clear that multiple groups, most prominently the LGBTQ community, were at risk from this new disease. The virus itself was discovered in 1983, and more and more was discovered about the etiological agent and the disease it caused5.
Reverse Transcriptase and Mutation
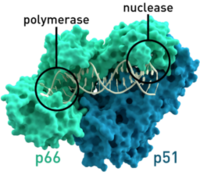
HIV relies on multiple proteins to replicate and propagate itself. One of these is reverse transcriptase, which transcribes RNA from the virus into DNA to be integrated into the genome of the host. This is a critical function for the replication of the virus, because HIV does not have the necessary translational and replication machinery, relying on the host to transcribe HIV’s genetic information along with the host information to create the proteins it needs to form progeny.
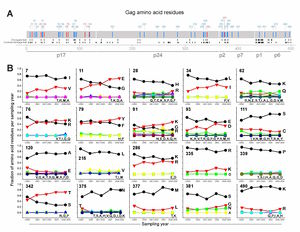
It is known that HIV’s reverse transcriptase is a “sloppy” enzyme. In the transcription from RNA to DNA, reverse transcriptase makes many mistakes, which leads to a high mutation rate. These mutations allow HIV to change rapidly and combat any host defenses that may arise, such as cytotoxic T lymphocytes produced by the immune system. As well, the mutation of the virus allows for fast evolution and the adaptation to the host, which may lead to higher virulence and pathogenicity. In a paper published in Science, a group showed that on average, the HIV genome encodes roughly 5-10 mutations per round of replication. This is roughly one error every 4000 nucleotides7. In contrast, it is estimated that the human genome, with its highly advanced and accurate repair mechanisms, encodes only one mutation in every 30 million base pairs20. While some mutations are deleterious and lead to decreased fitness, some increase fitness of viral progeny and propagate the disease. For instance, parts of HIV proteins that white blood cells recognize change quickly and evade the immune system in that way (Figure 2).
Drug Therapy and Drug Resistance
With the introduction of antiretroviral therapy, the prognosis for an HIV infection became markedly better. However, challenges remain in drug treatment, which can cost over $15,000 a year in the United States, representing up to 84% of HIV/AIDS treatment costs16.
HIV Life Cycle
Highly active antiretroviral therapy (HAART) works by targeting the virus at multiple points in its life cycle. HIV needs to attach to CD4 and CCR5 or CXCR4 on the cell surface. HIV then needs to fuse with the cell membrane to release its RNA into the cell, which is blocked by fusion inhibitors, which inhibit both binding and fusion. HIV then reverse transcribes its RNA into DNA, which is blocked by reverse transcriptase inhibitors. Nucleoside reverse transcription inhibitors work by acting as a nucleobase analog that can be incorporated into the growing chain, but terminate extension. Non-nucleoside reverse transcription inhibitors work by binding directly to the reverse transcriptase enzyme and inhibiting its function. The reverse transcribed DNA is then integrated into the genome, which can be blocked by integrase inhibitors. Once the DNA is integrated, the host transcribes and translates the DNA into RNA and protein. HIV’s proteins are unique in that they are translated as a single long peptide chain, which must then cleaved by proteases to form mature proteins. This is blocked by protease inhibitors. Once the necessary proteins and RNA made, the new virus is packaged and buds from the cell to infect a fresh host cell8.
Development of Resistance
Because HIV mutates quickly, HAART is comprised of a multi-drug cocktail. By administering multiple drugs, usually three, that target multiple points in HIV's life cycle, it minimizes the chances that HIV can develop resistance to all the drugs at once and create an escape mutant. However, if the levels of drug present in the body are not enough to completely suppress and block replication, but enough to exert a selection pressure on the viral population, then resistance can quickly develop. As well, if the drug regimen is not followed strictly every day, and is instead sporadic, then this offers opportunity for the virus to adapt and mutate.These resistant strains can be passed among people, much like antibiotic resistant bacteria9.
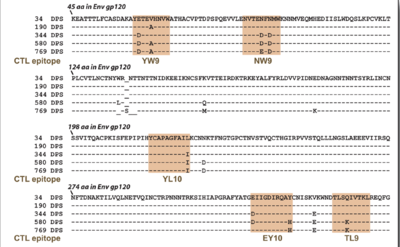
For instance, nucleoside and nucleotide reverse transcriptase inhibitors can be blocked by HIV through two types of mutations. The attached nucleotide or nucleoside can be removed through an ATP or pyrophosphate attack on the phosphodiester bond of the chain. This type of mutant is classified as thymidine analogue mutations. As well, the active site of the reverse transcriptase enzyme can be mutated to no longer fit the analogue bases, such as changing methionine at position 184 to valine. Another class of drug, the non-nucleoside reverse transcriptase inhibitor, works by binding to the active site of the enzyme. However, mutations can quickly accumulate which change the conformation and hydrophobicity of the binding pocket. Occasionally a single point mutation can markedly change the geometry and properties of the active site. This leads to a decreased affinity for the drug and a parallel increase in resistance to the compound9. There have been documented resistant strains to almost every non-nucleotide reverse transcriptase inhibitor (Figure 3). When the virus mutates and escape the control of the drug therapy, if a doctor notices that viral load increases despite HAART, then the drug regimen must be quickly changed in order to suppress further mutation and increased pathogenicity.
Epidemiology and the Future
Varying Responses to HIV
A person’s response to the infection is primarily dictated by his or her human leukocyte antigen locus and the major histocompatibility complex that this locus encodes for. A study by Payne et al. looks into the response that the virus takes when presented with an HLA type that is highly protective because escape mutants have modulated their behavior to adapt. The study compared populations that have been exposed to the virus for different periods of time: Botswana, where the virus has been established for a longer period of time, and South Africa. It suggests that virulence decreases with time as the virus becomes adapted to the human population and the various HLA subtypes, as there is less of a protective effect from previously protective HLA subtypes in Botswana, while the same HLA subtype in South Africa conferred more protection. The authors hypothesize that this is happening with antiretroviral therapy as well11.
A competing study, published by The Lancet HIV, examines a specific population in Europe, and reached a different conclusion. They found that virulence has actually increased, mainly based by the faster drop in CD4+ counts in HIV infected patients, the main way to diagnose the induction of AIDS12. However, the contradictory findings may be because of two different populations with two different genetic backgrounds and two different contexts that could change the response of the virus. It is important to note that there are many different responses to the disease and that findings may not be generalizable, due to many variables that can affect the expression and response to the virus.
Future of HIV Treatment
it is clear that the mutation and evolution of HIV poses challenges to the treatment and prevention of HIV infection.Development of new drug therapeutics is ongoing, as is research into vaccines and other prevention methods. A newly introduced method is PrEP, which gives HIV antiretrovirals, specifically the branded drug Truvada (emtricitabine/tenofovir) , to populations at high risk for infection. With these drugs circulating in the bloodstream, HIV does not have a chance to replicate and establish an infection[17].
Promising new therapies for HIV/AIDS includes genomic editing through the CRISPR-Cas9 system, which can specifically target specific loci in a genome and perform exquisitely precise editing by deletion, insertion, or a change of genetic information. The Δ32 mutation in CCR5, which is found in some European populations, changes the conformation of CCR5, a critical coreceptor needed for HIV entry into the cell. These people present as phenotypically normal, yet HIV cannot enter into their cells, so they are effectively immune to HIV infection. A striking example is the "Berlin patient." He was effectively cured of HIV when his treatment for leukemia was to ablate his immune system and receive a bone marrow transplant from a matched donor with this specific mutation14. This reconstituted his immune system with cells that were resistant to HIV infection, and to this day he has remained virus free. Therefore, if CRISPR-Cas9 can create this mutation in stem cells, creating a population of infection resistant hematopoietic progenitor cells and lymphocytes, this could render a person immune to infection.
Another finding that was published in Nature in 2015 by Gardner et al. was the announcement that a promising pseudo-vaccine had been created that showed protective responses in chimps against SIV challenges. When injected into muscle cells, the vector inserted DNA into the genome that, when expressed, would code for an engineered antibody-like protein. This protein mimicked CD4 and CCR5, which are crucial for HIV entry into CD4+ cells. These attach to HIV envelope proteins and prevent HIV from entering the cell15.
HIV is a formidable target that is constantly changing and evolving to the pressures that we bring to bear on this disease, taking advantages of opportunities to grow and spread. However, with many avenues of inquiry and research, we can hopefully one day find a treatment for HIV that does not simply slow down the progression of HIV/AIDS, but directly leads to a reversal of infection and a permanent cure.
References
1. "Global Statistics" (2014). AIDS.gov.
2. Zimmer, Carl (2015). "Two Strains of H.I.V. Cut Vastly Different Paths." New York Times.
5. “History of AIDS Up to 1986."(2014). Avert.org.
8. “The HIV Life Cycle.” (2013). AIDSinfo. National Institutes of Health.
9. Clavel, F. et al. (2004). “HIV drug resistance.” The New England Journal of Medicine 350: 1023-1035.
14. Ledford, Heidi (2009). "Stem-cell transplant wipes out HIV." Nature News.
17. "PrEP" (2015). Centers for Disease Control and Prevention.
Edited by Sam Du, a student of Suzanne Kern in BIOL168L (Microbiology) in The Keck Science Department of the Claremont Colleges Spring 2015.
