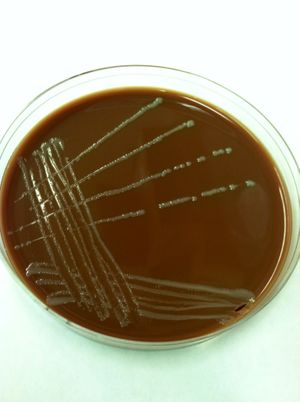
Francisella noatunensis
Classification
Domain: Bacteria
Phylum: Proteobacteria
Class: Gammaproteobacteria
Order: Thiotrichales
Family: Francisellaceae
Francisella noatunensis
|
NCBI: Taxonomy |
Description and Significance
Francisella noatunensis (F. noatunensis) are part of the family of Francisellaceae, which are aerobic, non-motile, coccobacilli gram-negative bacteria. Another feature of F. noatunensis is that they are facultative intracellular organisms in which they grow inside or outside of host cell. Being the cause of francisellosis, a fish disease that results in the development of granulomas, it has been a growing problem amongst fish farms and wild fish. There have been epidemics spanning across the globe affecting threeline grunt in Japan, tilapia in the Americas and Asia, hybrid striped bass in the Americas, Atlantic salmon in Chile, and Atlantic cod in Norway. With most of these epidemics affecting fish farms, the growing problem is resulting in a scramble to find out more about F. noatunensis and how it is causing the widespread problem. Recent research has suggested that there is no sign of zoonotic potential for humans. Which has been a concern as the closely related F. tularensis is a well-known human pathogen that causes tularemia [1].
Structure, Metabolism, and Life Cycle
F. noatunensis is a pathogenic capsulated coccobacilli bacterium that has recently been found to live in endosymbiosis with the protist Euplotes raikovi (E. raikovi) [1]. Due to the challenge of cultivation of Francisella in laboratory, specific medias have been developed. It has been found that the organisms requires cysteine and specifically grows well on cysteine heart agar with a low percentage of ovine blood incorporated. Another interesting feature of F. noatunensis, is the fact that they lack the production of cytochrome oxidase. This feature has been used to distinguish them as the notable fish pathogen [2]. With cultivation being somewhat difficult PCR techniques to identify their presence has been used with 16S rRNA and also detecting certain genes of their 1.85 Mb genome [2][3].
Like most intracellular bacterium they go through a specific life cycle. Individual F. noatunensis bacterium encapsulated by a phagosome first enters by undergoing phagocytosis into a host cell. Next the phagosomal escape stage is entered; this is where the bacterium is released into the cytosol. This is followed by cytosolic replication and ultimately cell death, when all the replicated F. noatunensis are released [4].
Ecology and Pathogenesis
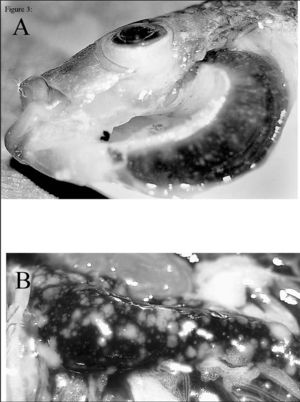
Seeing as F. noatunensis is a common fish pathogen causing francisellosis, it has mostly been found in to survive in fresh and marine water environments. With the recent discovery of it being found to live in the water dwelling protist E. raikovi, this would suggest a probable vector for them to exist before becoming pathogenic. Although little is still known about the pathogenesis of F. noatunensis, its found to be transmitted in contaminated waters. What is known is it causes granulomas in fish and eventually leads to high mortality rates among fish populations where contaminated water is found. Recent studies suggest water temperature plays a role in transmission, as the warmer the water at which the fish can survive, the higher frequency of disease being found. Also, since culture has been successful from Atlantic cod gut, this implies perhaps fecal-oral transmission is at play. The identification of F. noatunensis in fish eggs may additionally suggest vertical transmission of the disease. Overall, more research on the pathogenesis and transmission is being done to sort out these unknown mechanisms [1][2].
References
[1] Schrallhammer, M., Schweikert, M., Vallesi, A., Verni, F., and Petroni, G. (2011). Detection of a Novel Subspecies of Francisella noatunensis as Endosymbiont of the Ciliate Euplotes raikovi. Microb. Ecol. 61, 455–464. http://link.springer.com/article/10.1007/s00248-010-9772-9
[2] Colquhoun, D.J., and Duodu, S. (2011). Francisella infections in farmed and wild aquatic organisms. Vet. Res. 42, 47. http://www.veterinaryresearch.org/content/42/1/47
[3] Sjödin, A., Svensson, K., Ohrman, C., Ahlinder, J., Lindgren, P., Duodu, S., Johansson, A., Colquhoun, D.J., Larsson, P., and Forsman, M. (2012). Genome characterisation of the genus Francisella reveals insight into similar evolutionary paths in pathogens of mammals and fish. Bmc Genomics 13, 268. http://www.biomedcentral.com/1471-2164/13/268
[4] Chong, A., and Celli, J. (2010). The Francisella intracellular life cycle: toward molecular mechanisms of intracellular survival and proliferation. Front. Cell. Infect. Microbiol. - Closed Sect. 1, 138. http://www.frontiersin.org/Cellular_and_Infection_Microbiology_-_closed_section/10.3389/fmicb.2010.00138/abstract
Author
Page authored by Joseph Navratil, student of Mandy Brosnahan, Instructor at the University of Minnesota-Twin Cities, MICB 3301/3303: Biology of Microorganisms.