HIV Drug Resistance
Introduction
By Lillian Spetrino
HIV Structure and Life Cycle
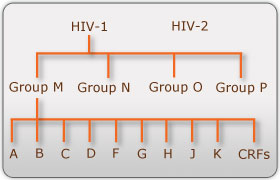
There are two known types of HIV – HIV type 1 and HIV type 2. Although both are transmitted through bodily fluids, HIV-2 is less pathogenic than HIV-1 and is thus less widely spread. There are four subtypes of HIV-1: the "major" group M, the "outlier" group O, group N, and group P. Within the major group are 9 clades (Figure 1) that are genetically distinct from one another and tend to be associated with certain geographical regions. Approximately 90% of HIV-1 infections belong to group M. It has been found that subtypes may have different infectivity levels and modes of transmission. Even though these subtypes are genetically distinct from one another, no change in effectiveness of antiretroviral drugs has been observed between subtypes (Advert).
HIV-1 is a spherical enveloped virus about 120 nm in diameter. The envelope is composed of a protein complex that controls membrane fusion with host CD4+ cells (Chan, 1998). The genome of HIV is comprised of two copies of (+) single stranded RNA. HIV is transmitted though bodily fluids. Sexual intercourse is the foremost manner of transmission, but is also common through intravenous drug users. Transmission is important to understand in order to prevent the transmission of resistant strains of HIV, which are more difficult to treat.
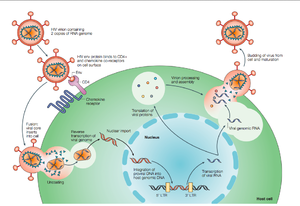
HIV enters the CD4+ T cell though tight binding to membrane proteins, causing the membranes to fuse and allows entry of the viral capsid in the host cell (Figure 2). Once inside the cell, HIV RNA and reverse transcriptase, integrase, ribonuclease, and protease are used to reverse transcribe the (+) RNA into cDNA, integrate the cDNA into the host cell genome, transcribe into mRNA, and translate and package into virions. These immature virions are transported to the plasma membrane where protease cleaves the gag polyproteins into the matrix, capsid, and nucleocapsid. Following, the mature virion buds off the host cell and is capable of infecting other cells. Replication of HIV is essential for its survival, and for developing resistance to HIV drugs.
HIV Therapy
Prior to 1996, HIV was considered a fatal illness. However the development of combination antiretroviral therapy has decreased HIV-associated morbidity and mortality (Panel on Antiretroviral Guidelines for Adults and Adolescents, 2014) and transformed HIV into what is now considered a manageable, albeit chronic, disease (Bangsberg, 2007). Antiretroviral therapy is highly effective in transmission prevention between sexual partners (Panel on Antiretroviral Guidelines for Adults and Adolescents, 2014). The treatment goals of antiretroviral therapy are to reduce HIV-associated illness and prolong the length and quality of life, repair and preserve immunologic function, suppress plasma HIV viral load, and block HIV transmission (Panel on Antiretroviral Guidelines for Adults and Adolescents, 2014). However, an undesired result of antiretroviral therapy are mutations that can lead to decreased drug vulnerability (Novak, 2005). Drug-resistance HIV patients have restricted treatment options and have been found to have reduced response to therapy (Novak, 2005).
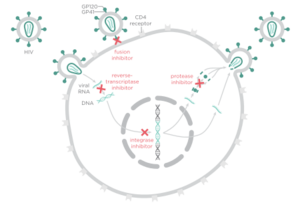
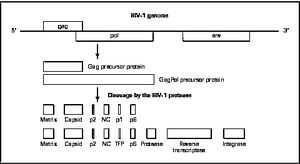
There are four major classes of HIV treatment drugs (Figure 3), targeted towards a specific aspect of HIV proliferation. One such class is nucleoside and nucleotide analogues. This drug terminates the DNA chain and inhibits reverse transcription of the viral RNA. Nonnucleoside reverse-transcriptase inhibitors are another type of HIV treatment drugs that bind and inhibit reverse transcriptase. Additionally, entry inhibitors block the entry of HIV virions into their target cells. Lastly, protease inhibitors target the viral enzyme protease. Protease cleaves the precursor proteins gag and gag-pol (Figure 5), which is important for viral maturation (Clavel, 2004). In order to become resistant to these drugs, the virus must constantly replicate in order to create mutations that confer resistance. In this sense, it is a constant struggle between the drugs preventing replication and the virus attempting to evade the drugs and develop resistance.
Resistance
HIV has a high rate of evolution (Smyth, 2012) and therefore abnormally elevated genetic diversity. This genetic variability is a result of the rapid replication cycle. The replication cycle of HIV has a generation time of about 2.5 days, producing between 1010 and 1012 new virions each day. This in combination with the high mutation rate (approximately 0.2 errors by reverse transcriptase per genome during each replication cycle) and the ability to frequently recombine (Rambaut, 2004), confers a high mutation rate. Modulation of HIV-1 mutation rate by reverse transcriptase can cause polymorphisms that alter replication fidelities resulting in drug resistance. The genetic diversity in the form of a viral quasispecies is associated with the failure of antiretroviral therapies due to drug resistance. Because of the nature of HIV-1 replication and the tendency for rapid selection, high-level drug resistance is a potential hurdle to overcome when treating HIV (Smyth, 2012).
Another potential cause of drug resistance in HIV is incomplete adherence. According to the World Health Organization, adherence to long term therapy is “the extent to which a person’s behavior – taking medication, following a diet, and/or executing lifestyle changes, corresponds with agreed recommendations from a health care provider” (World Health Organization, 2003). Incomplete adherence therefore is failure to follow such instructions from a health care professional and has often been thought to lead to drug resistance because at low adherence, replication is not as restricted, thus mutations conferring resistance are likely to arise. Recent data has suggested that each regiment class might have a special relationship between adherence and resistance (Bangsberg, 2007).
In order to avoid resistance, HIV-1 patients are treated using combination therapy. Combination therapy uses three drugs from at least two classes (Panel on Antiretroviral Guidelines for Adults and Adolescents, 2014) in order to increase the barrier to resistance. Combination therapy helps to decrease the likelihood of resistance because it reduces HIV replication at multiple steps in viral replication and thus reduces the likelihood that a resistance mutation will occur. Since resistance requires multiple mutations to develop against all the drugs in the treatment, combination therapy is efficient at defending against resistance. However, combination therapy is unable to wholly eliminate resistance to antiretroviral drugs because highly active antiretroviral therapy cannot completely overpower viral replication (Smyth, 2012), which opens up the unfortunate possibility to become resistant to all drugs in the cocktail.
The goal of HIV treatment is to reduce the amount of virus to indiscernible levels, or less than 50 copies of the virus per mL, ideally 24 weeks after beginning combination therapy. If viral load is found to be higher than 200 copies per mL, further testing must be considered to determine the cause of the higher levels. If the virus has become resistant, for example, the virus would have no trouble replicating and viral load would be high. Another way to test how well the treatment is working is to use CD4+ cell counts. If the number of CD4+ cells is less than 50-100 cells per mL after a year (with 25% fluxuation), it may be indicative of insufficient antiretroviral therapy effectiveness. If there are low viral counts accompanied with low CD4+ cell counts, this is termed “immunologic nonresponse” or “immunologic failure” (Panel on Antiretroviral Guidelines for Adults and Adolescents, 2014 and Gazzola, 2009) and is cause for reevaluation of treatment.
HIV-1 Protease
Structure and Function
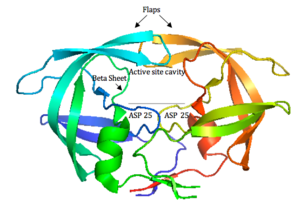
HIV-1 protease (Figure 4) is a homodimer composed of two identical subunits of 99 amino acids. Great plasticity and polymorphisms have been observed in 1/3 of the amino acids (Wensing, 2010). Also included are two molecular “flaps” that move up to 7 Å when the enzyme is charged with a substrate (Miller, 1989). This movement of the flaps is what allows the substrate to enter or depart the cleft (Marseveen). The active site is located between identical subunits and has the classic Asp-Thr-Gly sequence found in many aspartic proteases. More specifically, the catalytic activity of the enzyme is found between the Asp25 residues.
Mature infectious virus particles are generated when HIV protease cleaves viral Gag and GagPol precursor proteins (Figure 5). Gag encodes capsid, nucleocapsid, and matrix proteins (Slonczewski, 2013), while GagPol encodes RT, integrase, and protease (Slonczewski, 2013). Mechanistically, water acts as the nucleophile in combination with aspartic acid to hydrolyze the peptide bond (Jaskólski, 1991). This cleavage occurs during or shortly after the assembled viral particles are expelled from host cells and are therefore only essential for viral maturation, but are not required for viral production (Wensing, 2010).
Protease Inhibitors
Protease inhibition prevents the selective cleavage of viral polyproteins important for viral maturation, instead forming non-infections viral particles. Even though protease inhibitors reduce viral replication, some residual viral activity persists, which can lead to mutations that lower binding affinity of the protease to the inhibitor. Such mutations can occur both inside the active site and outside the binding cleft.
One issue with protease inhibitors is that many of them have poor oral bioavailability or short half lives, which causes patients to require large amounts of the drug more frequently. This leads to a potential problem with adherence which can lead to resistance. In addition, large volumes of the protease inhibitor can have short term toxic effects, such as gastrointestinal side effects, rash, renal colic, dry skin, and paronychia. Most concerning for patients was the resulting fat redistribution linked with protease inhibitors in 2-83% of patients on protease inhibitor therapy, further increasing the likelihood of inadequate adherence (Walmsley, 2007).
In order to combat these issues with protease inhibitors, there was a move towards using nonnucleoside reverse transcriptase inhibitors. However this came with its own set of issues: pregnant women cannot use nonnucleoside reverse transcriptase inhibitors because they can cause birth defects and cause hepatotoxicity in women with CD4 counts less than or equal to 250 cells per mm3 and men with CD4 counts less than or equal to 400 cells per mm3. Therefore, the lower pill burden was incapable of overcoming the negative side effects of nonnucleoside reverse transcriptase inhibitors, especially when the potential to become resistant to both nonnucleoside reverse transcriptase inhibitors and nucleoside reverse transcriptase inhibitors is possible (Walmsley, 2007). Therefore, improved protease inhibitors with more accessible dosing schedules was required. The solution scientists came up with was to “boost” the protease inhibitors.
Protease inhibitors are metabolized in the liver by CYP3A isoenzymes. Therefore if cytochrome p450 is induced or inhibited, metabolism of protease inhibitors may be impacted (Panel on Antiretroviral Guidelines for Adults and Adolescents, 2014). This is the reason why many protease inhibitors are often boosted with ritonavir, a CYP inhibitor. Boosted protease inhibitors significantly increase the levels of protease inhibitors in the blood plasma. However, this pharmacologic boosting can potentially raise protease inhibitor levels too much, causing deleterious effects, particularly gastrointestinal and hypertriglyceridemia (Walmsley, 2007).
There are some advantages and disadvantages to using protease inhibitors in HIV treatment. They are advantageous in the sense that it is more difficult to genetically develop resistance than it is for other HIV treatment drugs such as non-nucleoside reverse transcriptase inhibitors. Another advantage is that at the time of treatment failure, protease inhibitor resistance is uncommon when the protease inhibitor is ritonavir-boosted (Panel on Antiretroviral Guidelines for Adults and Adolescents, 2014). Unfortunately, there are also disadvantages to using protease inhibitors. Some metabolic complications that can arise are dyslipidemia, insulin resistance, and hepatotoxicity as well as gastrointestinal adverse effects. Also, there is a potential drug interaction is with CYP3A4 inhibitors and substrates, this interaction being more pronounced with ritonavir-based regiments (Walmsley, 2007).
Protease Inhibitor Resistance
Protease inhibitors were initially designed to mimic the scissile bonds of natural protease inhibitor substrates. However, these protease inhibitors resulted in high levels of resistance and numerous side effects. The next step then was to develop "second generation" protease inhibitors that favored enthalpic and entropic contributions (Kožíšek et al, 2014).
Viral reverse transcriptase lacks proofreading activity, and thus led to viral variants which retain mutations (Maarseveen, 2006). As the number of protease inhibitors has increased, it is important to be able to delineate how each protease inhibitor mutation impacts resistance to other protease inhibitors (Rhee, 2010). Estimated prevalence for protease inhibitor resistance is 0.7% (Novak, 2005).
High level resistance requires several mutations, and these mutations must reduce the ability of the virus to replicate. Boosted protease inhibitor regimens have been found to be more effective at decreasing viral replication than unboosted protease inhibitor regimens. In addition, it was discovered that protease mutations that have the largest influence on protease inhibitor vulnerability are those that are not polymorphic in the absence of protease inhibitor therapy (Rhee, 2010).
Resistance Mechanism
Protease inhibitor resistance usually begins with mutations likely to reduce binding affinity with the target, typically in the active site, thereby reducing replication capacity (Bangsberg, 2007). It is considered to be a stepwise process (Wensing, 2010) whereby an alteration in the active site of the protease is altered in some way. Typically, there is a substitution in the substrate-binding cleft of the viral protease resulting in enlargement of the catalytic site. This enlargement leads to decreased binding to the inhibitor (and therefore causing a decrease in drug sensitivity) as well as the natural substrate, which leads to decreased viral replication. These can be considered primary, or major, mutations. Secondary, or minor, mutations may also occur, however these do not typically have significant effect on resistance phenotype alone. Nonetheless, secondary mutations in conjunction with major mutations will improve replication of viruses.
Another potential mechanism of resistance is through an amino acid insertion. Amino acid insertions ranging from 1 to 6 amino acids have been observed during protease inhibitor-based antiretroviral therapy. Such insertions may lead to decrease in protease inhibitor susceptibility and slightly improve viral replication, but only contribute to resistance to protease inhibitors when in combination with other mutations in protease or Gag protein. Insertions have a positive correlation with protease mutations associated with protease inhibitor resistance whose usage has increased in recent years (atazanavir, lopinavir, amprenavir, and tipranavir) (Wensing, 2010)
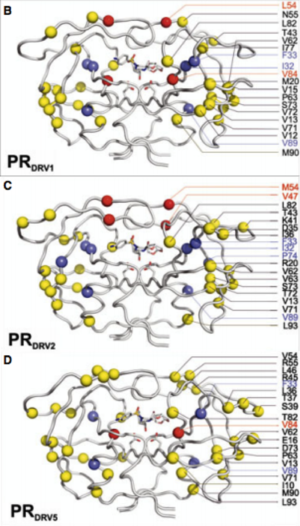
Another site of resistance is in the molecular flaps associated with entry of the substrate. These flaps can either be open, semi-open, or closed. MD and X-ray crystallography indicate that without a substrate, HIV-1 protease favors the semi open conformation, whereas inhibitor binding induces a closed flap conformation. In a study by Huang et al (2014), on a mutated sample of HIV-1 subtype B (PR5), they used pulsed electron double resonance, double electron-electron resonance, and NMR 15N relaxation measurements in order to characterize polymorphisms found in the mutated HIV-1 protease. They discovered that there was a novel conformation in the flap region that they hypothesize to be a curled-open state or an asymmetric configuration when interacting with inhibitors (Figure 6). The curled-open state is similar to the open flap conformation missing from HIV-1 subtype B protease. This new confirmation results from the removal of a salt bridge in the enzyme which destabilizes the fold of the flap, thus forming the new confirmation. These changes in backbone dynamics are suggested to be involved in lowering the effects of protease inhibitors (Huang et al., 2014).
Other mechanisms of resistance include mutations in the Gag protein in or near the protease cleavage sites. This is typically in response to the altered substrate-binding cleft of the mutant drug-resistant viral protease (Wensing, 2010). Mutations in viral protease in combination with mutations in Gag protein increase protease inhibitor resistance (Wensing, 2010). Another alternative pathway is a substitution in the Gag NC/p1 cleave site without any alterations in the viral PR sequence that causes protease inhibitor resistance leading to increased gag processing (Wensing, 2010). Furthermore, cleavage site (CS) mutations have been found to accumulate in protease inhibitor-resistant HIV isolates, correlating with the presences of three or more protease inhibitor resistance mutations. Mon-cleavage site mutations in Gag were also found in the frame-shift regulating site were also observed to be associated with resistance. In one experiment by Knops, 25% of HIV isolates from patients failing protease inhibitor treatment without protease mutations possessed Gag mutations associated with protease inhibitor resistance (Knops,2012). These Gag mutations could potentially indicate the evolution of protease inhibitor resistance, even without mutations in the protease enzyme.
Specific Drug Resistance
Darunavir

Darunavir is a second generation HIV protease inhibitor that is very active against wild type and resistant HIV strains. However, amino acid mutations in 21 positions that have been reported in patient samples. Kožíšek et al. studied clinically derived mutant protease inhibitors resistant to Darunavir in an effort to understand the mechanism behind the resistance development. Samples were clinically derived from a series of six mutant darunavir resistant HIV proteases. Amino acid sequences of protease variants were analyzed and compared to the wild type sequence. Up to 20 mutations in the resistant proteases decreased affinity of darunavir up to 13,000 fold (Figure 7).
Also investigated was the binding affinity of Darunavir to the mutant protease variants and the wild type enzyme (Figure 8) and found that these mutations in the resistant proteases decreased binding affinity of Darunavir. Interestingly, protease "DRV3" did not have any mutations associated with resistance. Instead, it carried the I50L mutation, which should confer higher darunavir susceptibility. This sample have a relatively lower binding affinity of darunavir than the other protease variants. However, there exists an unfavorable entropic factor which causes the resistance observation (Kožíšek et al, 2014)
Nelfinavir
Nelfinavir is a protease inhibitor that binds tightly to protease in the active site. The drug cannot be boosted, meaning it cannot be paired with ritonavir to pharmacologically increase amounts of nelfinavir in the blood by inhibiting metabolism of nelfinavir by cytochrome p450 (Walmsey, 2004). Because of this, nelfinavir is less successful at reducing viral load than protease inhibitors that can be boosted.
The protease mutation D30N is the mutation associated with resistance in nelfinavir treated patients. However, it only confers moderate levels of phenotypic resistance, suggesting that in order to develop resistance to nelfinavir, the patient must be exposed to high levels of the drug. This is in opposition to the non-adherence theory of resistance. In Bangsberg’s experiment, clinically derived patient samples were treated in vitro at variable concentrations of nelfinavir in order to simulate different levels of adherence. It was observed that resistance mutations were only observed after relatively high levels of drug exposure. In fact, 85% to 95% adherence was found to be the optimum adherence range for the selection of nelfinavir resistance (Bangsberg, 2007).
Lopinavir (Bangsberg, 2007)
Lopinavir is a protease inhibitor that is ritonavir-boosted. Drug resistance is uncommon in this case when patients are treatment-naïve, most likely because more than one mutation is required to generate resistance. These mutations must reduce the reproductive capabilities of the virus. Administration of ritonavir-boosted lopinavir increases’ the amount of lopinavir in the bloodstream, which makes it hard for the virus to be replicated, even when the patient is non-adherent. This is likely the case for other boosted protease inhibitor combinations.
Pharmacogenomic variation
Differing polymorphisms in CYP3A and CYP2C genes may lead to different adherence-resistance relationships, which may influence drug resistance.
Resistance Management
The first signs of drug resistance is an increase in plasma viremia (“a marker of antiviral activity and real-time phenotypic evaluation of viral isolates for the emergence of resistance” [Davey]) above the normal limits. Once it is determined that the increased levels are due to resistance, choosing therapy for treatment is a nuanced interaction between the patient and the health care professional. Typically, an almost monotherapy regimen is developed using newly developed agents that have novel viral targets. (Cortez, 2011)
One option discussed by Dr. Scholmo Staszewski, from the University of Frankfurt in Germany, is the role of “double PI” boosting as a potential therapy for patients with drug resistance, but remains skeptical and suggests further studies into drug combinations before use clinically.
Conclusion
HIV is highly genetically variable due to the nature of its replication. Because of this genetic flexibility, mutations may arise which confer resistance to certain HIV treatment drugs. One such class of drugs are protease inhibitors. Protease inhibitors are designed to bind to the active site of the viral enzyme protease to competitively inhibit protease from processing the Gag and Gal/Pol protein precursors, preventing the complete maturation of the HIV virion. However, resistance to these inhibitors can occur, specifically in the active site of the protease. Such drug specific mutations, referred to as primary mutations, are typically near the active site and in flap domains that provide access to the active site. Also theoretically present are secondary mutations, which, alone do not typically confer resistance to therapy but can increase resistance if present in conjunction with primary mutations. Resistance to protease inhibitors is increasingly rare if the HIV management therapy is combined with ritonavir boosted treatments. Ritonavir is a cytochrome p450 inhibitor that is used with protease inhibitors to pharmacologically boost protease inhibitor levels because cytochrome p450 can metabolize many protease inhibitors.
References
1. Bangsberg, David R., Deanna L. Kroetz, and Steven G. Deeks. "Adherence-resistance Relationships to Combination HIV Antiretroviral Therapy." Current HIV/AIDS Reports 4.2 (2007): 65-72. Print. DOI 10.1007/s11904-007-0010-0
2. Bonhoeffer S, Chappey C, Parkin NT, Whitcomb JM, Petropoulos CJ. (2004). Evidence for positive epistasis in HIV-1. Science 2004 306(5701):1547–1550. Erratum in: Science. 2008 May 9;320(5877):746. doi:10.1126/science.1101786 PMID 15567861
3. Chan, D. C., & Kim, P. S. (1998). HIV entry and its inhibition. Cell, 93(5), 681-684. doi:http://dx.doi.org/10.1016/S0092-8674(00)81430-0
4. Clavel, F., & Hance, A. J. (2004). HIV drug resistance. N Engl J Med, 350(10), 1023-1035. doi:10.1056/NEJMra025195
5. Cortez KJ, Maldarelli F. Clinical management of HIV drug resistance. Viruses.2011;3(4):347–378. doi: 10.3390/v3040347
6. Gazzola L, Tincati C, Bellistrì GM, et al. (2009) The absence of CD4+ T cell count recovery despite receipt of virologically suppressive highly active antiretroviral therapy: clinical risk, immunological gaps, and therapeutic options. Clin Infect Dis 2009; 48: 328–337. doi: 10.1086/595851
7. "HIV Strains: Types, Groups and Subtypes." HIV and AIDS Information and Resources. Advert, 2011. Web. 02 May 2014.
8. Huang, X et al., (2014). The role of select subtype polymorphisms on HIV-1 protease conformational sampling and dynamics. J. Biol. Chem. 10.1074/jbc.M114.571836
9. Jaskólski M, Tomasselli AG, Sawyer TK, Staples DG, Heinrikson RL, Schneider J, Kent SB, Wlodawer A (February 1991). "Structure at 2.5-A resolution of chemically synthesized human immunodeficiency virus type 1 protease complexed with a hydroxyethylene-based inhibitor". Biochemistry 30 (6): 1600–9.doi:10.1021/bi00220a023. PMID 1993177.
10. Knops, E., Brakier-Gingras, L., Schulter, E., Pfister, H., Kaiser, R., & Verheyen, J. (2012). Mutational patterns in the frameshift-regulating site of HIV-1 selected by protease inhibitors. Medical Microbiology and Immunology, 201(2), 213-218. [1]
11. Kožíšek, M., et al., (2014). Thermodynamic and structural analysis of HIV protease resistance to darunavir ? analysis of heavily mutated patient-derived HIV-1 proteases. FEBS Journal, 281(7), 1834-1847. doi:10.1111/febs.12743
12. Miller M, Schneider J, Sathyanarayana BK, Toth MV, Marshall GR, Clawson L, Selk L, Kent SB, Wlodawer A (December 1989)."Structure of complex of synthetic HIV-1 protease with a substrate-based inhibitor at 2.3 A resolution". Science 246(4934): 1149–52. doi:10.1126/science.2686029.
13. Novak, R. M., Chen, L., MacArthur, R. D., Baxter, J. D., Huppler Hullsiek, K., Peng, G.,Terry Beirn Community Programs for Clinical Research on AIDS 058 Study Team. (2005). Prevalence of antiretroviral drug resistance mutations in chronically HIV-infected, treatment-naive patients: Implications for routine resistance screening before initiation of antiretroviral therapy. Clinical Infectious Diseases : An Official Publication of the Infectious Diseases Society of America, 40(3), 468-474. doi:CID33817
14. Perryman AL, Lin JH, McCammon JA (April 2004). "HIV-1 protease molecular dynamics of a wild-type and of the V82F/I84V mutant: possible contributions to drug resistance and a potential new target site for drugs". Protein Sci. 13 (4): 1108–23. doi:10.1110/ps.03468904. PMC 2280056. PMID 15044738.
15. Slonczewski, J., Foster, W. Microbiology: An Evolving Science, 3rd Edition. W. W. Norton & Company, 2013-10-17. VitalBook ;file.
16. Smyth, R. P., Davenport, M. P., & Mak, J. (2012). The origin of genetic diversity in HIV-1. Virus Research, 169(2), 415-429. doi: 10.1016/j.virusres.2012.06.015.
17. Panel on Antiretroviral Guidelines for Adults and Adolescents (2014). Guidelines for the use of antiretroviral agents in HIV-1-infected adults and adolescents. Department of Health and Human Services. Available at http://aidsinfo.nih.gov/ContentFiles/AdultandAdolescentGL.pdf.
18. Rambaut, Andrew, David Posada, Keith A. Crandall, and Edward C. Holmes. "The Causes and Consequences of HIV Evolution." Nature Reviews Genetics 5.1 (2004): 52-61. doi:10.1038/nrg1246
19. Rhee SY, Taylor J, Fessel WJ, Kaufman D, Towner W, Troia P,Ruane P, Hellinger J, Shirvani V, Zolopa A, Shafer RW. 2010. HIV-1 protease mutations and protease inhibitor cross-resistance. Antimicrob. Agents Chemother. 54:4253–4261 doi: 10.1128/AAC.00574-10.
20. van Maarseveen N, Boucher C. 2006. Resistance to protease inhibitors. In: Geretti AM, editor. Antiretroviral Resistance in Clinical Practice. London: Mediscript; 2006. Chapter 3.Available from: [2]
21. Walmsley, Sharon. 2004. Overview of Boosted Protease Inhibitors from the Seventh international Congress on Drug Therapy in HIV Infection – Canadian Thought-Leader Perspective From Sharon Walmsey, MD, MSc [3]
22. Walmsley, S. (2007). Protease inhibitor-based regimens for HIV therapy: Safety and efficacy. Journal of Acquired Immune Deficiency Syndromes (1999), 45 Suppl 1, S5-13; quiz S28-31. doi:10.1097/QAI.0b013e3180600709 [doi]
23. Wensing, A. M. J., van Maarseveen, N. M., & Nijhuis, M. (2010). Fifteen years of HIV protease inhibitors: Raising the barrier to resistance. Antiviral Research, 85(1), 59-74. doi:http://dx.doi.org/10.1016/j.antiviral.2009.10.003
doi:http://dx.doi.org/10.1016/j.antiviral.2009.10.003
24. WHO. Adherence to long-term therapies; evidence for action. 2003. [4]