Inoviridae: Difference between revisions
Slonczewski (talk | contribs) |
|||
| (15 intermediate revisions by 3 users not shown) | |||
| Line 1: | Line 1: | ||
{{Curated}} | |||
{{Viral Biorealm Family}} | |||
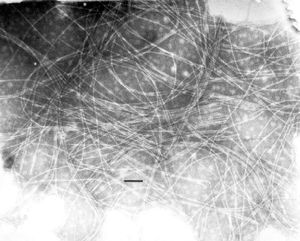
[[Image:inoviridae.jpg|thumb|right|(EM of Thermus phage H75, a member of Inoviridae. From [http://www.ncbi.nlm.nih.gov/ICTVdb/Images/Ackerman/Phages/Inovirid/773-08.htm ICTVdB-Picture Gallery]; Courtesy Dr. Hans Ackermann]] | |||
==Baltimore Classification== | ==Baltimore Classification== | ||
| Line 7: | Line 12: | ||
===Genera=== | ===Genera=== | ||
Inovirus, Plectovirus | ''Inovirus'', ''Plectovirus'' | ||
==Description and Significance== | ==Description and Significance== | ||
Inoviridae viruses are non-enveloped | Inoviridae viruses are non-enveloped bacteriophages that infect E. coli by a slow-release life cycle. | ||
==Genome Structure== | ==Genome Structure== | ||
The genome of inoviridae is not segmented and contains a molecule of circular, positive-sense, single-stranded DNA. The complete genome is 4400-8500 nucleotides long. (source: [http://www.ncbi.nlm.nih.gov/ICTVdb/ICTVdB/00.035.htm ICTV dB Descriptions]) | |||
==Virion Structure of an Inoviridae== | |||
The virions of inoviridae consist of a capsid that is not enveloped. The capsid is elongated (filamentous) and shows helical symmetry. The capsid is straight to flexuous with a length of 85-280 nm or 760-1950 nm, and a width of 10-16 nm or 6-8 nm. Frequently morphologically aberrant forms have also been observed and they are of abnormal length. (source: [http://www.ncbi.nlm.nih.gov/ICTVdb/ICTVdB/00.035.htm ICTV dB Descriptions]) | |||
==Reproduction Cycle of an Inoviridae in a Host Cell== | ==Reproduction Cycle of an Inoviridae in a Host Cell== | ||
The genome of inoviridae replicates either independently by the rolling-circle mechanism like free plasmids or with the chromosome if the viral genome becomes integrated. There are five steps in the normal productive infectious cycle: phage adsorption and uptake of the infecting ssDNA circle, conversion of the ssDNA circle to a parental replicative form (RF) by host cell enzymes, semiconservative RF replication initiated by a viral endonuclease, synthesis of progeny ssDNA sequestered by ssDNA binding protein, and the membrane based-assembly process that extrudes progeny virions into the medium. | |||
The genes of inovirus are closely spaced and several genes are translated from overlapping reading frames or from alternate starts in the same frame. Intergenic regions contain the complementary and viral strand replication origins and DNA packaging signals. Phage adsorption involves specific interactions between another domain of gp3 with other host membrane proteins. The ssDNA is converted to a supercoiled dsDNA replicative form (RF) by cellular enzymes. Phage DNA replication begins when the viral endonuclease gp2 expressed from parental RF nicks this RF at a specific, high-symmetry site. Progeny RF produced via ssDNA intermediates in rolling-circle replication become templates for further RF replication and further mRNA synthesis. Gp10 and gp5 can downregulate the nicking activity of gp2. When sufficient gp5 is made, complementary strand synthesis is blocked and complexes of gp5 and progeny viral ssDNA accumulate. Assembly is initiated at the membrane by concerted interactions of gp7, gp9, gp1, and gp11 and a specific packaging signal in a hairpin on the ss DNA in the gp5 complex. Assembly proceeds at the inner membrane where about 1500 subunits of gp5 are displaced by 2700 subunits of gp8. Both gp1 and gp11 appear to be involved in this transfer of the DNA from gp5-ssDNA complexes into the assmbling virion. Gp1 may also function in the formation of adhesion zones between the inner and outer membranes by interacting with outer membrane pores formed by subunits of viral protein gp4. Assembly of virions is completed by addition of gp6 and gp3. There are notable execptions to this overall pathway. Lysogenic strains encode integrases, and viral sequences, partial or complete, are found integrated at several chromosomal sites. In Cf1t and Cf16 variants of Cf1c, the first inovirus lysogens characterized, the gene for site specific integration shows no homology with any Ff genes. The Vibrio phage CTX is another species with lysogenic phase and its genome encodes the two subunits of cholera toxin. The toxin genes become highly expressed and the toxin is released upon conversion of the lysogen to the nonlytic productive cycle. The <i>Plectovirus</i> species <i>Acholeplasma phage</i> MV-L51 (L51) has been shown to be similar to this scheme with respect to DNA replication pathways, and virions are assembled at the membranes as they are released into the medium without lysis. This is presumably true for all species in the genus replicating as indeoendent plasmids and producing virus by extrusion. The genomes of two viruses (L1 and SpV1) have both been found to be integrated into the host chromosome at one or more sites. | |||
==Viral Ecology & Pathology== | ==Viral Ecology & Pathology== | ||
| Line 26: | Line 37: | ||
==References== | ==References== | ||
Day, L.A., & Maniloff, J.; "Inoviridae"; <u>The Seventh Report of the International Committee on Taxonomy of Viruses</u>; 2000 | |||
[http://www.ncbi.nlm.nih.gov/ICTVdb/ICTVdB/00.035.htm ICTV dB Descriptions] | |||
Latest revision as of 01:15, 3 July 2011
A Viral Biorealm page on the family Inoviridae
Baltimore Classification
Higher order taxa
Viruses; ssDNA viruses; Inoviridae
Genera
Inovirus, Plectovirus
Description and Significance
Inoviridae viruses are non-enveloped bacteriophages that infect E. coli by a slow-release life cycle.
Genome Structure
The genome of inoviridae is not segmented and contains a molecule of circular, positive-sense, single-stranded DNA. The complete genome is 4400-8500 nucleotides long. (source: ICTV dB Descriptions)
Virion Structure of an Inoviridae
The virions of inoviridae consist of a capsid that is not enveloped. The capsid is elongated (filamentous) and shows helical symmetry. The capsid is straight to flexuous with a length of 85-280 nm or 760-1950 nm, and a width of 10-16 nm or 6-8 nm. Frequently morphologically aberrant forms have also been observed and they are of abnormal length. (source: ICTV dB Descriptions)
Reproduction Cycle of an Inoviridae in a Host Cell
The genome of inoviridae replicates either independently by the rolling-circle mechanism like free plasmids or with the chromosome if the viral genome becomes integrated. There are five steps in the normal productive infectious cycle: phage adsorption and uptake of the infecting ssDNA circle, conversion of the ssDNA circle to a parental replicative form (RF) by host cell enzymes, semiconservative RF replication initiated by a viral endonuclease, synthesis of progeny ssDNA sequestered by ssDNA binding protein, and the membrane based-assembly process that extrudes progeny virions into the medium.
The genes of inovirus are closely spaced and several genes are translated from overlapping reading frames or from alternate starts in the same frame. Intergenic regions contain the complementary and viral strand replication origins and DNA packaging signals. Phage adsorption involves specific interactions between another domain of gp3 with other host membrane proteins. The ssDNA is converted to a supercoiled dsDNA replicative form (RF) by cellular enzymes. Phage DNA replication begins when the viral endonuclease gp2 expressed from parental RF nicks this RF at a specific, high-symmetry site. Progeny RF produced via ssDNA intermediates in rolling-circle replication become templates for further RF replication and further mRNA synthesis. Gp10 and gp5 can downregulate the nicking activity of gp2. When sufficient gp5 is made, complementary strand synthesis is blocked and complexes of gp5 and progeny viral ssDNA accumulate. Assembly is initiated at the membrane by concerted interactions of gp7, gp9, gp1, and gp11 and a specific packaging signal in a hairpin on the ss DNA in the gp5 complex. Assembly proceeds at the inner membrane where about 1500 subunits of gp5 are displaced by 2700 subunits of gp8. Both gp1 and gp11 appear to be involved in this transfer of the DNA from gp5-ssDNA complexes into the assmbling virion. Gp1 may also function in the formation of adhesion zones between the inner and outer membranes by interacting with outer membrane pores formed by subunits of viral protein gp4. Assembly of virions is completed by addition of gp6 and gp3. There are notable execptions to this overall pathway. Lysogenic strains encode integrases, and viral sequences, partial or complete, are found integrated at several chromosomal sites. In Cf1t and Cf16 variants of Cf1c, the first inovirus lysogens characterized, the gene for site specific integration shows no homology with any Ff genes. The Vibrio phage CTX is another species with lysogenic phase and its genome encodes the two subunits of cholera toxin. The toxin genes become highly expressed and the toxin is released upon conversion of the lysogen to the nonlytic productive cycle. The Plectovirus species Acholeplasma phage MV-L51 (L51) has been shown to be similar to this scheme with respect to DNA replication pathways, and virions are assembled at the membranes as they are released into the medium without lysis. This is presumably true for all species in the genus replicating as indeoendent plasmids and producing virus by extrusion. The genomes of two viruses (L1 and SpV1) have both been found to be integrated into the host chromosome at one or more sites.
Viral Ecology & Pathology
References
Day, L.A., & Maniloff, J.; "Inoviridae"; The Seventh Report of the International Committee on Taxonomy of Viruses; 2000
