Lactobacillus plantarum
Classification
Domain : Bacteria
Phylum : Firmicutes
Class : Bacilli
Order : Lactobacillales
Family : Lactobacillaceae
Genus : Lactobacillus
Species : plantarum
Lactobacillus plantarum (8)
Description and Significance
L. plantarum is a gram positive bacteria that is found in a variety of niches. These niches include dairy, meat, and many vegetable fermentations, it is also found in the human gastrointestinal tract. It is a facultative heterofermentative lactic acid bacterium that utilizes an extensive range of fermentatable carbon sources. Lactic acid bacteria are Gram-positive and they are non-spore forming, fermentative bacteria that grow anaerobically. The main function of these bacteria is the fermentative conversion of sugars present in raw materials into lactic acid. L. plantarum also produces anti-microbial peptides and exopolysaccharides. It has the ability to maintain a pH gradient between the inside and outside of the cell in the presence of large amounts of acetate or lactate. (3) L. plantarum is one of the most common microbes used as a silage innocculant. Silage is a fermented fodder that can be fed to ruminants or used as a biofuel feedstock for anaerobic digesters. (4)
L. plantarumis currently being explored to convert lignocellulosic biomass to biofuel and bioproducts. Current research into this idea are looking at a strain of L. plantarum which has certain genes inactivated to eliminate undesirable fermentation products. (7)L. plantarum is also able to degrade cassava raw starch. Its ability to degrade raw starch is useful because it could potentially be used as a starter in certain traditional fermentation processes. (4) There are also potential uses for L. plantarum to be used in treatments of certain wastewater due to its ability to degrade phenolic compounds, such as those in olive mill wastewaters. (1)
The ability of the this microbe to adapt and thrive in a range of environments, its ability and capacity to be genetically manipulated, as well as its ability to ferment and degrade different materials makes L. plantarum a very interesting and important bacteria to study.
Genome Structure
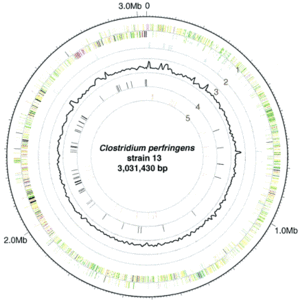
L. plantarum has one of the largest genomes among lactic acid bacteria. In its circular chromosome it contains 3,308,274 base pairs. The genome was sequenced by using whole genome sequencing as assembly approach. The overall GC content of its chromosome is 44.5%, the plasmids tend to have a lower percent GC content. Putative biological functions have been given to 2,120 of the predicted proteins. One particular interesting region of the chromosome is the 213-kb region from 3,072,500 – 3,28,500, which encodes proteins for sugar transport, metabolism, and regulation. This region has a lower percent CG content (41.5%), leading researchers to believe that many of these genes have been acquired by horizontal gene transfer. (5)
There are three plasmids contained in the L. plantarum chromosome, pWCFS101, pWCFS102, and pWCFS103. The plasmid sizes are as follows: pWCFS101 contains 1,917 bp, pWCFS102 contains 2,365 bp, and pWCFS103 contains 36,069 bp. Plasmid pWCFS101 is believed to contain replication proteins. Plasmid pWCFS102 is believed to contain replication proteins as well as proteins that function as copy number controls. Plasmid pWCFS103 contains genes that are predicted to be involved in arsenate and/or aresenite resistance as well as cadmium resistance; it also has genes that are believed to encode replication proteins, resolvases, DNA-damage-inducible proteins, and oxidases. L. plantarum contains two apparently complete prophage genomes, as well as some prophage remnants. (6)
The L. plantarum chromosome reveals that this microbe has a major focus on carbon catabolism. The sequence of its chromosome also supports its extreme flexibility, versatility, and ability to adapt to different environmental conditions. (5)
Cell Structure, Metabolism and Life Cycle
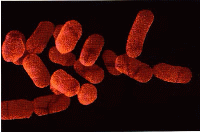
Lactobacillus plantarum is a rod shaped structure with the ends being rounded. This is a gram positive bacteria meaning there is a high concentration of peptidoglycan in the cell wall and lacks an outer cell membrane.
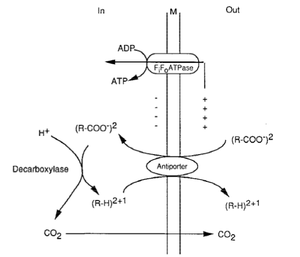
L. plantarum is a facultative heterofermentative lactobacilli microorganism. This means that the organism takes carbon from sugars and the byproduct is either alcohol or lactic acid. This process happens in an aerotolerant environment meaning that oxygen is not present. L. plantarum is also a manganese dependent process which uses manganese as a pseudo catalase which lowers oxygen concentration which is favorable to the aerotolerant environment.
Lactobacillus plantarum gains energy in various forms which include sugar and pyruvate. During the degradation of sugar carbon is released and becomes a source of energy for the lactobacillus plantarum.
When L. plantarum is exhibiting a pyruvate metabolism it is similar to homolactic fermentation. This happens when growth occurs on glucose which is degraded pyruvate though an EMP pathway. Once the pyruvate is formed it is converted to d and l-lactate though sterospecific lactate dehydrogenase enzymes (2).
Ecology and Pathogenesis
L. plantarum can be found in many different environments, most commonly it is isolated from plant material and the human gastrointestinal tract. (3) Researchers believe that the sequence of the L. plantarum genome has certain features that allow this microbe to be versatile and adaptive to different environments. (5) This versatility allows L. platarum isolates to be found in human saliva, fermenting dairy products, plant material, silage, and even certain waste waters. It gains its energy through the fermentative conversion of sugars to lactic acid, as long as is able to go through this process, most environments will allow the growth of this microbe.
Recently L. plantarum has been identified as a probiotic. Probiotics are non-pathogenic microorganisms that can have a positive impact on human health when they are digested. They are becoming a very popular dietary supplement to many people, especially those who have gastrointestinal problems. (9) In this case, L. plantarum can be considered a human symbiont. When the probiotics are injested regularly it is possible that the composition of microflora in the intestinal tract can be maipulated. This manipulation may allow an improvement of microbe balance, stabilization of digestive enzyme patterns, and immunomodulation by activating and regulating mucosa-associated and systemic immune system responses. The microflora foudn in the intestinal tract are thought to provide protection from pathogens. Some companies currently sell bottles containing L. plantarum as a probiotic to help with intestinal problems including IBS and IBD, stating that these bacteria help to "Balance the Intestinal Ecosystem." http://www.customprobiotics.com/about_probiotics.htm
Habitat; symbiosis; biogeochemical significance; contributions to environment.
References
Author
Page authored by Stephanie LaHaye and Jason McIntyre, student of Prof. Jay Lennon at Michigan State University.

