Limnohabitans (genus)
Classification
Domain: Bacteria
Phylum: Proteobacteria
Class: Betaproteobacteria
Order: Burkholderiales
Family: Comamonadaceae
Genus: Limnohabitans
Description
The genus Limnohabitans belongs to the class Betaproteobacteria, which is well known for its morphological and physiological diversity, as well as its high abundance compared to other bacterial groups in freshwater lakes. Due to its abundance and easy ability to culture, Betaproteobacteria best-studied group in freshwater lakes.
Betaproteobacteria has been divided into seven lineages - betI, betII, betIII, betIV, betV, betVI, and betVII. Members of the genus Limnohabitans are represented in the betI lineage, which have recently been isolated in pure culture and accurately described. There are four Limnohabitans species within the betI lineage - L. curvus, L. planktonicus, L. parvus, L. australis. These four species can be distinguished their substrate utilization characteristics, 16S rRNA gene sequences and extent of genomic DNA-DNA hybridization.
FUN FACT: The Limnohabitans species itself is relatively new in the research world, with first documented studies occurring around 2009 and much information is still in the process of being discovered!
Habitat
Limnohabitans live in freshwater environments in habitats that have a neutral pH. They are sensitive to acidity and salinity, so their occurrence in ocean or acidic environments is negligible. In freshwater lakes, they inhabit the neuston, the epilimnion, and the hypolimnion levels, due to their capability to live in both oxic and anoxic environments. Their high abundance makes them a prime target for protist grazing and flagellates.
Ecology and Significance
The Betaproteobacteria in freshwater lakes are fast-growing, nutrient-dependent, and have a size-dependent vulnerability to grazing pressure. The betI and betII lineages are the most abundant and widely distributed lineages in freshwater ecosystems. Recent isolations of various members of genus Limnohabitans, which belong to the lineage betI, have assisted in the understanding of freshwater bacterial roles in their ecosystems.
A current study is being conducted, using Limnohabitans as a model for testing responses of natural flagellate communities to different bacterial food quality. They are discovering that with Limnohabitans, there are prey-related difference in flagellate growth parameters, which influences the community composition of flagellates.
Also, due to their relatively recent discovery, they are being used as a model in another study to test their role in carbon flow to higher trophic levels. This study has found evidence that suggests that Limnohabitans has a key role in carbon flow from algal-derived substrates to the plankton grazer food chain.
Morphology and Genome Structure
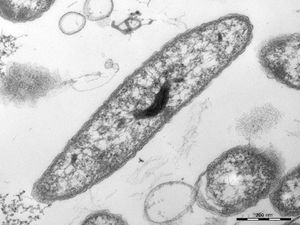
Limnohabitans have varying morphologies and structures. Isolated strains have been described as coccoid, ovoid, bacillus (curved and short-rod) or solenoid. Cell sizes range from 0.4 um in diameter of cocci and up to 5 um in length of curved bacilli.
According to GenBank, isolates from a population of Limnohabitans, although clearly distinct from currently recognized species, have not yet been characterized using traditional methods or validly published. Therefore, genome sequences are not yet fully available. However, the Pathosystems Resource Integration Center (PATRIC) has obtained potential genome data for Limnohabitans sp. Rim28 and sp. Rim 47. For example, the Limnohabitans sp. Rim 28 had 3389652bp and a GC content of 58.5%.

Metabolism
Limnohabitans demonstrates high growth rates and metabolic versatility, dependent on their relationship with algal-derived organic substances. Growth of L. parvus and L. panktonicus on algal exudates as a sole dissolved organic carbon source (DOC) has recently been confirmed. The photolysis of this dissolved organic matter creates products that are suspected to be an important additional substrate source for Limnohabitan bacteria.
References
https://microbewiki.kenyon.edu/index.php/Limnohabitans_planktonicus Razak, Shairah, and Malachi Schuster. "Limnohabitans planktonicus." MicrobeWiki. N.p., n.d. Web. 12 May 2015.
http://aem.asm.org/content/77/20/7307.full Simek, Karel. "Alga-Derived Substrates Select for Distinct Betaproteobacterial Lineages and Contribute to Niche Separation in Limnohabitans Strains." Applied and Environmental Microbiology. N.p., 2011. Web. 12 May 2015.
http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3063352/ Newton, Ryan. "A Guide to the Natural History of Freshwater Lake Bacteria." NCBI. N.p., 2011. Web. 12 May 2015.
http://journals.plos.org/plosone/article?id=10.1371/journal.pone.0058527 Jezbera, Jan. "Patterns of Limnohabitans Microdiversity across a Large Set of Freshwater Habitats as Revealed by Reverse Line Blot Hybridization." PLOS One. N.p., 12 Mar. 2013. Web. 12 May 2015.
http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3591437/ Kasalický, Vojtěch. "The Diversity of the Limnohabitans Genus, an Important Group of Freshwater Bacterioplankton, by Characterization of 35 Isolated Strains." NCBI. PLOS One, 7 Mar. 2013. Web. 12 May 2015.
http://kbe.prf.jcu.cz/en/projekt/genus-limnohabitans-important-freshwater-bacterial-group-serves-model-testing-responses Šimek, Karel. "Genus Limnohabitans - an important freshwater bacterial group serves as a model for testing responses of natural flagellate communities to different bacterial food quality." Katedra Biologie Ekosystemu. N.p., 2013. Web. 12 May 2015
http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3721123/ Simek, Karel. "Differential freshwater flagellate community response to bacterial food quality with a focus on Limnohabitans bacteria." NCBI. The ISME Journal, 7 Aug. 2013. Web. 12 May 2015.
http://www.ncbi.nlm.nih.gov/genome/?term=Limnohabitans "Limnohabitans: Genome Assembly Report." NCBI. N.p., 2012. Web. 12 May 2015.
https://www.patricbrc.org/portal/portal/patric/GenomeList?cType=taxon&cId=665874&dataSource=&displayMode=&pk= "Limnohabitans Genome." Pathosystems Integration Resource Center. N.p., 2012. Web. 12 May 2015.
http://ijs.sgmjournals.org/content/60/6/1358.abstract?sid=df28eb96-e60d-48a9-8db7-9f9af637f1e3 Hahn, Martin. "Limnohabitans curvus gen. nov., sp. nov., a planktonic bacterium isolated from a freshwater lake." International Journal of Systematic and Evolutionary Microbiology. N.p., 11 Aug. 2009. Web. 12 May 2015.
Figures
Author
Page authored by Taylor Wagner, student of Prof. Katherine Mcmahon at University of Wisconsin - Madison.